Pythonで体験するベイズ推論 PyMCによるMCMC入門 第4章 写経
目次
始めに
この記事では、Pythonで体験するベイズ推論 PyMCによるMCMC入門
の第4章を写経した記録をまとめます。前回はこちらPythonで体験するベイズ推論 PyMCによるMCMC入門 第3章 (3.2全部) 写経。 実行はJupyter Labにて実行しています。
写経は以下のようにまとめました。
写経ではありますが、関数を作成し、コードの再実行をやりやすいようにした部分があります。
より良いと思われるコードを考えた場合は書き換えてコメントを添えるようにし、変更点をなるべく明示するようにしてあります。
個人的に気になった点のメモを残すようにしてあります。同じような疑問を持った方の助けになれば幸いです。
以前書いたコードと同じようなコード(例えばグラフの描写等)は効率化のために飛ばしているところもあります。
記事内で使用するモジュールなどは一番最初に宣言するようにしてあります。
Pythonで体験するベイズ推論 PyMCによるMCMC入門を読んでいている際のちょっとした正誤表代わりになればと思います。
この記事で使用する主なモジュール、設定
この記事では、主に以下のモジュールや設定を使用しています。import prawを実行する前に、pip install praw==3.6.0を実行してprawモジュールをインストールしておかないとエラーが出ます。
from os import makedirs from urllib.request import urlretrieve import praw import pymc as pm from IPython.core.pylabtools import figsize import matplotlib.pyplot as plt
4 偉大な定理、登場
4.3 サンプルサイズが小さいという災い
4.3.3 例題 : Redditコメントをソートする
makedirs("data", exist_ok=True) urlretrieve("https://git.io/vXtX2", "top_showerthoughts_submissions.py")
Output:
('top_showerthoughts_submissions.py',
<http.client.HTTPMessage at 0x1ce88b88f60>)
%run top_showerthoughts_submissions.py 2 print("Title of submission:") print(top_post) # top_postはtop_showerthoughts_submissions.pyを実行した際に代入される。型は文字列である。
Output:
Title of submission:
April Fools Day is proof the Purge wouldn’t work. Despite having a whole day for shitposting, there is still shitposting the rest of the year.
n_comments = len(contents) comments = np.random.randint(n_comments, size=4) print("Some Comments (out of %d total) \n----------" % n_comments) for i in comments: print('"' + contents[i] + '"') print("upvotes/downvotes: ", votes[i, :], "\n")
Output:
Some Comments (out of 2 total)
----------
"That guy who is always kicking robots in those Boston Dynamics videos is gonna have a really bad day when the machines take over."
upvotes/downvotes: [44 2]
"That guy who is always kicking robots in those Boston Dynamics videos is gonna have a really bad day when the machines take over."
upvotes/downvotes: [44 2]
"That guy who is always kicking robots in those Boston Dynamics videos is gonna have a really bad day when the machines take over."
upvotes/downvotes: [44 2]
"Shaving cream is like a progress bar of how much you've got left to shave."
upvotes/downvotes: [40 3]
メモ
どうやら、まさかのコメントが2個しかないということが判明した。時間がたってしまって変化があったのかも・・・シンプルに
comments = [0, 1]
として進めます。
comments = [0, 1] for i in comments: print('"' + contents[i] + '"') print("upvotes/downvotes: ", votes[i, :], "\n")
Output:
"That guy who is always kicking robots in those Boston Dynamics videos is gonna have a really bad day when the machines take over."
upvotes/downvotes: [44 2]
"Shaving cream is like a progress bar of how much you've got left to shave."
upvotes/downvotes: [40 3]
def posterior_upvote_ratio(upvotes, downvotes, samples=20000): N = upvotes + downvotes upvote_ratio = pm.Uniform("upvote_ratio", 0, 1) observations = pm.Binomial("obs", N, upvote_ratio, value=upvotes, observed=True) map_ = pm.MAP([upvote_ratio, observations]).fit() mcmc = pm.MCMC([upvote_ratio, observations]) mcmc.sample(samples, samples / 4) return mcmc.trace("upvote_ratio")[:]
plt.rcParams['font.size'] = 17
figsize(22, 16) colors = ["#348ABD", "#A60628", "#7A68A6", "#467821", "#CF4457"] posteriors = [] for i in range(len(comments)): j = comments[i] label = f'({votes[j, 0]} up:{votes[j, 1]} down)\n{contents[j][:50]}...' posteriors.append(posterior_upvote_ratio(votes[j, 0], votes[j, 1])) plt.hist(posteriors[i], bins=18, density=True, alpha=.9, histtype="step", color=colors[i % 5], lw=3, label=label) plt.hist(posteriors[i], bins=18, density=True, alpha=.2, histtype="stepfilled", color=colors[i], lw=3) plt.legend(loc="upper left") plt.xlim(0, 1) plt.ylabel("Density") plt.xlabel("Probability of upvote") plt.title("Posterior distributions of upvote ratios " "on differennt comments")
Output:
[-----------------100%-----------------] 20000 of 20000 complete in 1.0 sec
Text(0.5, 1.0, 'Posterior distributions of upvote ratios on differennt comments')

N = posteriors[0].shape[0] lower_limits = [] for i in range(len(comments)): j = comments[i] label = f'({votes[j, 0]} up:{votes[j, 1]} down)\n{contents[j][:50]}...' plt.hist(posteriors[i], bins=20, density=True, alpha=.9, histtype="step", color=colors[i], lw=3, label=label) plt.hist(posteriors[i], bins=20, density=True, alpha=.2, histtype="stepfilled", color=colors[i], lw=3) v = np.sort(posteriors[i])[int(0.05 * N)] plt.vlines(v, 0, 10, color=colors[i], linestyles="--", linewidths=3) lower_limits.append(v) plt.legend(loc="upper left") plt.xlabel("Probability of upvote") plt.ylabel("Density") plt.title("Posterior distributions of upvote ratios " "on different comments") order = np.argsort(-np.array(lower_limits)) print(order, lower_limits)
Output:
[0 1] [0.87004917925665515, 0.82966075938026218]

4.3.5 でも計算が遅すぎる!
def intervals(u, d): a = 1. + u b = 1. + d mu = a / (a + b) std_err = 1.65 * np.sqrt((a * b) / ((a + b)**2 * (a + b + 1.))) return (mu, std_err) posterior_mean, std_err = intervals(votes[:, 0], votes[:, 1]) lb = posterior_mean - std_err print(f"Approximate lower bounds:\n{lb}, \n") order = np.argsort(-lb) ordered_contents = [] for i in order: ordered_contents.append(contents[i]) print(votes[i, 0], votes[i, 1], contents[i]) print("-----------------")
Output:
Approximate lower bounds:
[ 0.88044266 0.84187791],
44 2 That guy who is always kicking robots in those Boston Dynamics videos is gonna have a really bad day when the machines take over.
-----------------
40 3 Shaving cream is like a progress bar of how much you've got left to shave.
-----------------
r_order = order[::-1] plt.errorbar(posterior_mean[r_order], np.arange(len(r_order)), xerr=std_err[r_order], capsize=0, fmt="o", color="#7A68A6") plt.xlim(0.3, 1) plt.yticks(np.arange(len(r_order) - 1, -1, -1), map(lambda x: x[:30].replace("\n", ""), ordered_contents))
Output:
([<matplotlib.axis.YTick at 0x2730d10c0b8>,
<matplotlib.axis.YTick at 0x2730caaaf28>],
<a list of 2 Text yticklabel objects>)

メモ
map(lambda x: x[:30].replace("\n", ""),ordered_contents)はordered_contentsの各文字列の先頭30文字だけ抜き出し、改行を置換した要素を返すイテレータを生成します。
for i in map(lambda x: x[:30].replace("\n", ""),ordered_contents): print(i)
Output:
That guy who is always kicking
Shaving cream is like a progre
一般的に、mapは第一引数の関数を第二引数の各値に適用させたイテレータを返す関数です。
for i in map(lambda x: 2*x, range(10)): print(i)
Output:
0
2
4
6
8
10
12
14
16
18
メモ
今更だが、np.arangeの説明を読んでみた。rangeとほぼ同じような引数なんですね。
np.arange??(クリックすると展開されます)
Docstring:
arange([start,] stop[, step,], dtype=None)
Return evenly spaced values within a given interval.
Values are generated within the half-open interval ``[start, stop)``
(in other words, the interval including `start` but excluding `stop`).
For integer arguments the function is equivalent to the Python built-in
`range <http://docs.python.org/lib/built-in-funcs.html>`_ function,
but returns an ndarray rather than a list.
When using a non-integer step, such as 0.1, the results will often not
be consistent. It is better to use ``linspace`` for these cases.
Parameters
----------
start : number, optional
Start of interval. The interval includes this value. The default
start value is 0.
stop : number
End of interval. The interval does not include this value, except
in some cases where `step` is not an integer and floating point
round-off affects the length of `out`.
step : number, optional
Spacing between values. For any output `out`, this is the distance
between two adjacent values, ``out[i+1] - out[i]``. The default
step size is 1. If `step` is specified, `start` must also be given.
dtype : dtype
The type of the output array. If `dtype` is not given, infer the data
type from the other input arguments.
Returns
-------
arange : ndarray
Array of evenly spaced values.
For floating point arguments, the length of the result is
``ceil((stop - start)/step)``. Because of floating point overflow,
this rule may result in the last element of `out` being greater
than `stop`.
See Also
--------
linspace : Evenly spaced numbers with careful handling of endpoints.
ogrid: Arrays of evenly spaced numbers in N-dimensions.
mgrid: Grid-shaped arrays of evenly spaced numbers in N-dimensions.
Examples
--------
>>> np.arange(3)
array([0, 1, 2])
>>> np.arange(3.0)
array([ 0., 1., 2.])
>>> np.arange(3,7)
array([3, 4, 5, 6])
>>> np.arange(3,7,2)
array([3, 5])
Type: builtin_function_or_method
今回の写経は以上です。 ここまで読んでいただきありがとうございました。
Pythonで体験するベイズ推論 PyMCによるMCMC入門 第3章 (3.2全部) 写経
目次
始めに
この記事では、Pythonで体験するベイズ推論 PyMCによるMCMC入門
の第3章の3.2を写経した記録をまとめます。前回はこちらPythonで体験するベイズ推論 PyMCによるMCMC入門 第3章 (3.1全部) 写経。 実行はJupyter Labにて実行しています。
写経は以下のようにまとめました。
写経ではありますが、関数を作成し、コードの再実行をやりやすいようにした部分があります。
より良いと思われるコードを考えた場合は書き換えてコメントを添えるようにし、変更点をなるべく明示するようにしてあります。
個人的に気になった点のメモを残すようにしてあります。同じような疑問を持った方の助けになれば幸いです。
以前書いたコードと同じようなコード(例えばグラフの描写等)は効率化のために飛ばしているところもあります。
Pythonで体験するベイズ推論 PyMCによるMCMC入門を読んでいている際のちょっとした正誤表代わりになればと思います。
この記事で使用する主なモジュール、設定
この記事では、主に以下のモジュールや設定を使用しています。
from IPython.core.pylabtools import figsize from matplotlib import pyplot as plt import numpy as np import pymc as pm from pymc.Matplot import plot as mcplot
3 MCMCの中をのぞいてみよう
3.2 自己相関
figsize(25, 8) plt.rcParams['font.size'] = 17 x_t = pm.rnormal(0, 1, 200) x_t[0] = 0 y_t = np.zeros(200) for i in range(1, 200): y_t[i] = pm.rnormal(y_t[i - 1], 1) plt.plot(y_t, label="$y_t$", lw=3) plt.plot(x_t, label="$x_t$", lw=3) plt.xlabel("Time, $t$") plt.ylabel("Value") plt.title("Two different series of random values") plt.legend()
Output:
<matplotlib.legend.Legend at 0x27d4f39ecc0>

def autocorr(x): result = np.correlate(x, x, mode='full') result = result / np.max(result) # 通常の割り算だと、結果が浮動小数点になり、arrayのインデックスの引数に渡すと警告が出る。 #なので、//により商を求めるようにしておいた。 return result[result.size // 2:] colors = ["#348ABD", "#A60628", "#7A68A6"] x = np.arange(1, 200) plt.bar(x, autocorr(y_t)[1:], width=1, label="$y_t$", edgecolor=colors[0], color=colors[0]) plt.bar(x, autocorr(x_t)[1:], width=1, label="$x_t$", edgecolor=colors[1], color=colors[1]) plt.legend(title="autocorrelation") plt.ylabel("Measured correlation \n" "between $y_n$ and $y_{t-k}$.") plt.xlabel("$k$ (lag)") plt.title("Autocorrelation plot of $y_t$ and $x_t$ " "for differing $k$ lags")
Output:
Text(0.5, 1.0, 'Autocorrelation plot of $y_t$ and $x_t$ for differing $k$ lags')

3.2.2 間引き処理
max_x = 200 // 3 + 1 x = np.arange(1, max_x) plt.bar(x, autocorr(y_t)[1:max_x], edgecolor=colors[0], label="no thinning", color=colors[0], width=1) plt.bar(x, autocorr(y_t[::2])[1:max_x], edgecolor=colors[1], label="keeping every 2nd sample", color=colors[1], width=1) plt.bar(x, autocorr(y_t[::3])[1:max_x], edgecolor=colors[2], label="keeping every 3rd sample", color=colors[2], width=1) plt.autoscale(tight=True) plt.legend(title="Autocorrelation plot for $y_t$", loc="lower left") plt.ylabel("Measured correlation \n" "between $y_t$ and $y_{t-k}$.") plt.xlabel("$k$ (lag)") plt.title("Autocorrelation of $y_t$ (no thinning versus thinning) " "at differing $k$ lags")
Output:
Text(0.5, 1.0, 'Autocorrelation of $y_t$ (no thinning versus thinning) at differing $k$ lags')

p = pm.Uniform("p", 0., 1.) assignment = pm.Categorical("assignment", [p, 1-p], size=data.shape[0]) taus = 1.0 / pm.Uniform("stds", lower=0, upper=100, size=2) ** 2 # テキストを読んだ感じ、ここはupper=100なんじゃないんかな? centers = pm.Normal("centers", [120, 190], [0.01, 0.01], size=2) @pm.deterministic def center_i(assignment=assignment, centers=centers): return centers[assignment] @pm.deterministic def tau_i(assignment=assignment, taus=taus): return taus[assignment] observations = pm.Normal("obs", center_i, tau_i, value=data, observed=True) # これ、どこで使うんだ? model = pm.Model([p, assignment, taus, centers]) mcmc = pm.MCMC(model)
mcmc.sample(25000, 0, 10)
Output:
[-----------------100%-----------------] 25000 of 25000 complete in 9.7 sec
figsize(25, 8) plt.rcParams['font.size'] = 10 mcplot(mcmc.trace("centers", 2), common_scale=False)
Output:
Plotting centers_0
Plotting centers_1

今回の写経は以上です。 ここまで読んでいただきありがとうございました。
Pythonで体験するベイズ推論 PyMCによるMCMC入門 第3章 (3.1全部) 写経
目次
始めに
この記事では、Pythonで体験するベイズ推論 PyMCによるMCMC入門
の第3章の3.1を写経した記録をまとめます。前回はこちらPythonで体験するベイズ推論 PyMCによるMCMC入門 第2章 (2.2.6から2章の終わりまで) 写経。 実行はJupyter Labにて実行しています。
写経は以下のようにまとめました。
写経ではありますが、関数を作成し、コードの再実行をやりやすいようにした部分があります。
より良いと思われるコードを考えた場合は書き換えてコメントを添えるようにし、変更点をなるべく明示するようにしてあります。
個人的に気になった点のメモを残すようにしてあります。同じような疑問を持った方の助けになれば幸いです。
以前書いたコードと同じようなコード(例えばグラフの描写等)は効率化のために飛ばしているところもあります。
Pythonで体験するベイズ推論 PyMCによるMCMC入門を読んでいている際のちょっとした正誤表代わりになればと思います。
この記事で使用する主なモジュール、設定
この記事では、主に以下のモジュールや設定を使用しています。
import scipy.stats as stats import matplotlib as mpl from matplotlib import pyplot as plt %matplotlib inline from IPython.core.pylabtools import figsize import numpy as np from mpl_toolkits.mplot3d import Axes3D import pprint from os import makedirs from urllib.request import urlretrieve import pymc as pm
3 MCMCの中をのぞいてみよう
3.1 山あり谷あり、分布の地形
figsize(25, 8) plt.rcParams['font.size'] = 10 jet = plt.cm.jet fig = plt.figure() x = y = np.linspace(0, 5, 100) X, Y = np.meshgrid(x, y) plt.subplot(121) uni_x = stats.uniform.pdf(x, loc=0, scale=5) uni_y = stats.uniform.pdf(y, loc=0, scale=5) M = np.dot(uni_x[:, None], uni_y[None, :]) im = plt.imshow(M, interpolation='none', origin='lower', cmap=jet, vmax=1, vmin=-.15, extent=(0, 5, 0, 5)) plt.xlim(0, 5) plt.ylim(0, 5) plt.title("Overhead view of landscape formed by " "Uniform priors") ax = fig.add_subplot(122, projection='3d') ax.plot_surface(X, Y, M, cmap=plt.cm.jet, vmax=1, vmin=-0.15) ax.view_init(azim=390) ax.set_xlabel('Value of $p_1$') ax.set_ylabel('Value of $p_2$') ax.set_zlabel('Dencity') plt.title("Alternate view of landscape formed by " "Uniform priors")
Output:
Text(0.5, 0.92, 'Alternate view of landscape formed by Uniform priors')

メモ
np.meshgridは以下のような二つの行列を返しています。
np.meshgrid(x, y)
Output:
[array([[ 0. , 0.05050505, 0.1010101 , ..., 4.8989899 ,
4.94949495, 5. ],
[ 0. , 0.05050505, 0.1010101 , ..., 4.8989899 ,
4.94949495, 5. ],
[ 0. , 0.05050505, 0.1010101 , ..., 4.8989899 ,
4.94949495, 5. ],
...,
[ 0. , 0.05050505, 0.1010101 , ..., 4.8989899 ,
4.94949495, 5. ],
[ 0. , 0.05050505, 0.1010101 , ..., 4.8989899 ,
4.94949495, 5. ],
[ 0. , 0.05050505, 0.1010101 , ..., 4.8989899 ,
4.94949495, 5. ]]),
array([[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0.05050505, 0.05050505, 0.05050505, ..., 0.05050505,
0.05050505, 0.05050505],
[ 0.1010101 , 0.1010101 , 0.1010101 , ..., 0.1010101 ,
0.1010101 , 0.1010101 ],
...,
[ 4.8989899 , 4.8989899 , 4.8989899 , ..., 4.8989899 ,
4.8989899 , 4.8989899 ],
[ 4.94949495, 4.94949495, 4.94949495, ..., 4.94949495,
4.94949495, 4.94949495],
[ 5. , 5. , 5. , ..., 5. ,
5. , 5. ]])]
一つ目は、各行がnp.linspace(0, 5, 100)となっている100行100列の行列で、二つ目はその転置となっています。np.meshgridについては、等高線の描写などと相性が良いとdocstringに書いてありました。
print(np.meshgrid.__doc__) (クリックすると展開されます)
Return coordinate matrices from coordinate vectors.
Make N-D coordinate arrays for vectorized evaluations of
N-D scalar/vector fields over N-D grids, given
one-dimensional coordinate arrays x1, x2,..., xn.
.. versionchanged:: 1.9
1-D and 0-D cases are allowed.
Parameters
----------
x1, x2,..., xn : array_like
1-D arrays representing the coordinates of a grid.
indexing : {'xy', 'ij'}, optional
Cartesian ('xy', default) or matrix ('ij') indexing of output.
See Notes for more details.
.. versionadded:: 1.7.0
sparse : bool, optional
If True a sparse grid is returned in order to conserve memory.
Default is False.
.. versionadded:: 1.7.0
copy : bool, optional
If False, a view into the original arrays are returned in order to
conserve memory. Default is True. Please note that
``sparse=False, copy=False`` will likely return non-contiguous
arrays. Furthermore, more than one element of a broadcast array
may refer to a single memory location. If you need to write to the
arrays, make copies first.
.. versionadded:: 1.7.0
Returns
-------
X1, X2,..., XN : ndarray
For vectors `x1`, `x2`,..., 'xn' with lengths ``Ni=len(xi)`` ,
return ``(N1, N2, N3,...Nn)`` shaped arrays if indexing='ij'
or ``(N2, N1, N3,...Nn)`` shaped arrays if indexing='xy'
with the elements of `xi` repeated to fill the matrix along
the first dimension for `x1`, the second for `x2` and so on.
Notes
-----
This function supports both indexing conventions through the indexing
keyword argument. Giving the string 'ij' returns a meshgrid with
matrix indexing, while 'xy' returns a meshgrid with Cartesian indexing.
In the 2-D case with inputs of length M and N, the outputs are of shape
(N, M) for 'xy' indexing and (M, N) for 'ij' indexing. In the 3-D case
with inputs of length M, N and P, outputs are of shape (N, M, P) for
'xy' indexing and (M, N, P) for 'ij' indexing. The difference is
illustrated by the following code snippet::
xv, yv = meshgrid(x, y, sparse=False, indexing='ij')
for i in range(nx):
for j in range(ny):
# treat xv[i,j], yv[i,j]
xv, yv = meshgrid(x, y, sparse=False, indexing='xy')
for i in range(nx):
for j in range(ny):
# treat xv[j,i], yv[j,i]
In the 1-D and 0-D case, the indexing and sparse keywords have no effect.
See Also
--------
index_tricks.mgrid : Construct a multi-dimensional "meshgrid"
using indexing notation.
index_tricks.ogrid : Construct an open multi-dimensional "meshgrid"
using indexing notation.
Examples
--------
>>> nx, ny = (3, 2)
>>> x = np.linspace(0, 1, nx)
>>> y = np.linspace(0, 1, ny)
>>> xv, yv = meshgrid(x, y)
>>> xv
array([[ 0. , 0.5, 1. ],
[ 0. , 0.5, 1. ]])
>>> yv
array([[ 0., 0., 0.],
[ 1., 1., 1.]])
>>> xv, yv = meshgrid(x, y, sparse=True) # make sparse output arrays
>>> xv
array([[ 0. , 0.5, 1. ]])
>>> yv
array([[ 0.],
[ 1.]])
`meshgrid` is very useful to evaluate functions on a grid.
>>> x = np.arange(-5, 5, 0.1)
>>> y = np.arange(-5, 5, 0.1)
>>> xx, yy = meshgrid(x, y, sparse=True)
>>> z = np.sin(xx**2 + yy**2) / (xx**2 + yy**2)
>>> h = plt.contourf(x,y,z)
そして、ax.plot_surface(X, Y, M, cmap=plt.cm.jet, vmax=1, vmin=-0.15)では、XとYを座標としてMをプロットしているはずです。以下に、簡単な例を載せておきます。np.meshgridについては、等高線の描写などと相性が良いとdocstringに書いてありました。
figsize(12, 12) fig = plt.figure() ax_ = fig.add_subplot(111, projection='3d') x_ = np.array([0, 1]) y_ = np.array([0, 1]) X_, Y_ = np.meshgrid(x_, y_) M_ = np.array([[0, 1], [1, 2]]) print(f"X_:\n {X_}") print(f"Y_:\n {Y_}") ax_.plot_surface(X_, Y_, M_, cmap=plt.cm.jet, vmax=1, vmin=-0.15)
Output:
X_:
[[0 1]
[0 1]]
Y_:
[[0 0]
[1 1]]
<mpl_toolkits.mplot3d.art3d.Poly3DCollection at 0x1600073d710>

メモ
uni_x[:, None]は100行1列の行列、uni_y[None, :]は1行100列のベクトルになっています。従って、Mは100行100列の行列になります。
また、plt.imshow、plt.cm.jet、ax.plot_surface、ax.view_initの説明は以下のようになっています。plt.cm.jetはスカラーを色に変換する関数のような印象ですが、あまり理解できないですね・・・
print(plt.imshow.__doc__)(クリックすると展開されます)
Display an image, i.e. data on a 2D regular raster.
Parameters
----------
X : array-like or PIL image
The image data. Supported array shapes are:
- (M, N): an image with scalar data. The data is visualized
using a colormap.
- (M, N, 3): an image with RGB values (float or uint8).
- (M, N, 4): an image with RGBA values (float or uint8), i.e.
including transparency.
The first two dimensions (M, N) define the rows and columns of
the image.
The RGB(A) values should be in the range [0 .. 1] for floats or
[0 .. 255] for integers. Out-of-range values will be clipped to
these bounds.
cmap : str or `~matplotlib.colors.Colormap`, optional
A Colormap instance or registered colormap name. The colormap
maps scalar data to colors. It is ignored for RGB(A) data.
Defaults to :rc:`image.cmap`.
aspect : {'equal', 'auto'} or float, optional
Controls the aspect ratio of the axes. The aspect is of particular
relevance for images since it may distort the image, i.e. pixel
will not be square.
This parameter is a shortcut for explicitly calling
`.Axes.set_aspect`. See there for further details.
- 'equal': Ensures an aspect ratio of 1. Pixels will be square
(unless pixel sizes are explicitly made non-square in data
coordinates using *extent*).
- 'auto': The axes is kept fixed and the aspect is adjusted so
that the data fit in the axes. In general, this will result in
non-square pixels.
If not given, use :rc:`image.aspect` (default: 'equal').
interpolation : str, optional
The interpolation method used. If *None*
:rc:`image.interpolation` is used, which defaults to 'nearest'.
Supported values are 'none', 'nearest', 'bilinear', 'bicubic',
'spline16', 'spline36', 'hanning', 'hamming', 'hermite', 'kaiser',
'quadric', 'catrom', 'gaussian', 'bessel', 'mitchell', 'sinc',
'lanczos'.
If *interpolation* is 'none', then no interpolation is performed
on the Agg, ps and pdf backends. Other backends will fall back to
'nearest'.
See
:doc:`/gallery/images_contours_and_fields/interpolation_methods`
for an overview of the supported interpolation methods.
Some interpolation methods require an additional radius parameter,
which can be set by *filterrad*. Additionally, the antigrain image
resize filter is controlled by the parameter *filternorm*.
norm : `~matplotlib.colors.Normalize`, optional
If scalar data are used, the Normalize instance scales the
data values to the canonical colormap range [0,1] for mapping
to colors. By default, the data range is mapped to the
colorbar range using linear scaling. This parameter is ignored for
RGB(A) data.
vmin, vmax : scalar, optional
When using scalar data and no explicit *norm*, *vmin* and *vmax*
define the data range that the colormap covers. By default,
the colormap covers the complete value range of the supplied
data. *vmin*, *vmax* are ignored if the *norm* parameter is used.
alpha : scalar, optional
The alpha blending value, between 0 (transparent) and 1 (opaque).
This parameter is ignored for RGBA input data.
origin : {'upper', 'lower'}, optional
Place the [0,0] index of the array in the upper left or lower left
corner of the axes. The convention 'upper' is typically used for
matrices and images.
If not given, :rc:`image.origin` is used, defaulting to 'upper'.
Note that the vertical axes points upward for 'lower'
but downward for 'upper'.
extent : scalars (left, right, bottom, top), optional
The bounding box in data coordinates that the image will fill.
The image is stretched individually along x and y to fill the box.
The default extent is determined by the following conditions.
Pixels have unit size in data coordinates. Their centers are on
integer coordinates, and their center coordinates range from 0 to
columns-1 horizontally and from 0 to rows-1 vertically.
Note that the direction of the vertical axis and thus the default
values for top and bottom depend on *origin*:
- For ``origin == 'upper'`` the default is
``(-0.5, numcols-0.5, numrows-0.5, -0.5)``.
- For ``origin == 'lower'`` the default is
``(-0.5, numcols-0.5, -0.5, numrows-0.5)``.
See the example :doc:`/tutorials/intermediate/imshow_extent` for a
more detailed description.
shape : scalars (columns, rows), optional, default: None
For raw buffer images.
filternorm : bool, optional, default: True
A parameter for the antigrain image resize filter (see the
antigrain documentation). If *filternorm* is set, the filter
normalizes integer values and corrects the rounding errors. It
doesn't do anything with the source floating point values, it
corrects only integers according to the rule of 1.0 which means
that any sum of pixel weights must be equal to 1.0. So, the
filter function must produce a graph of the proper shape.
filterrad : float > 0, optional, default: 4.0
The filter radius for filters that have a radius parameter, i.e.
when interpolation is one of: 'sinc', 'lanczos' or 'blackman'.
resample : bool, optional
When *True*, use a full resampling method. When *False*, only
resample when the output image is larger than the input image.
url : str, optional
Set the url of the created `.AxesImage`. See `.Artist.set_url`.
Returns
-------
image : `~matplotlib.image.AxesImage`
Other Parameters
----------------
**kwargs : `~matplotlib.artist.Artist` properties
These parameters are passed on to the constructor of the
`.AxesImage` artist.
See also
--------
matshow : Plot a matrix or an array as an image.
Notes
-----
Unless *extent* is used, pixel centers will be located at integer
coordinates. In other words: the origin will coincide with the center
of pixel (0, 0).
There are two common representations for RGB images with an alpha
channel:
- Straight (unassociated) alpha: R, G, and B channels represent the
color of the pixel, disregarding its opacity.
- Premultiplied (associated) alpha: R, G, and B channels represent
the color of the pixel, adjusted for its opacity by multiplication.
`~matplotlib.pyplot.imshow` expects RGB images adopting the straight
(unassociated) alpha representation.
.. note::
In addition to the above described arguments, this function can take a
**data** keyword argument. If such a **data** argument is given, the
following arguments are replaced by **data[<arg>]**:
* All positional and all keyword arguments.
Objects passed as **data** must support item access (``data[<arg>]``) and
membership test (``<arg> in data``).
print(plt.cm.jet.__doc__)(クリックすると展開されます)
Colormap objects based on lookup tables using linear segments.
The lookup table is generated using linear interpolation for each
primary color, with the 0-1 domain divided into any number of
segments.
print(ax.plot_surface.__doc__)(クリックすると展開されます)
Create a surface plot.
By default it will be colored in shades of a solid color, but it also
supports color mapping by supplying the *cmap* argument.
.. note::
The *rcount* and *ccount* kwargs, which both default to 50,
determine the maximum number of samples used in each direction. If
the input data is larger, it will be downsampled (by slicing) to
these numbers of points.
Parameters
----------
X, Y, Z : 2d arrays
Data values.
rcount, ccount : int
Maximum number of samples used in each direction. If the input
data is larger, it will be downsampled (by slicing) to these
numbers of points. Defaults to 50.
.. versionadded:: 2.0
rstride, cstride : int
Downsampling stride in each direction. These arguments are
mutually exclusive with *rcount* and *ccount*. If only one of
*rstride* or *cstride* is set, the other defaults to 10.
'classic' mode uses a default of ``rstride = cstride = 10`` instead
of the new default of ``rcount = ccount = 50``.
color : color-like
Color of the surface patches.
cmap : Colormap
Colormap of the surface patches.
facecolors : array-like of colors.
Colors of each individual patch.
norm : Normalize
Normalization for the colormap.
vmin, vmax : float
Bounds for the normalization.
shade : bool
Whether to shade the face colors.
**kwargs :
Other arguments are forwarded to `.Poly3DCollection`.
ax.view_init??(クリックすると展開されます)
Signature: ax.view_init(elev=None, azim=None)
Source:
def view_init(self, elev=None, azim=None):
"""
Set the elevation and azimuth of the axes.
This can be used to rotate the axes programmatically.
'elev' stores the elevation angle in the z plane.
'azim' stores the azimuth angle in the x,y plane.
if elev or azim are None (default), then the initial value
is used which was specified in the :class:`Axes3D` constructor.
"""
self.dist = 10
if elev is None:
self.elev = self.initial_elev
else:
self.elev = elev
if azim is None:
self.azim = self.initial_azim
else:
self.azim = azim
File: c:\users\hoge\anaconda3\lib\site-packages\mpl_toolkits\mplot3d\axes3d.py
Type: method
figsize(25, 5) plt.rcParams['font.size'] = 13 fig = plt.figure() plt.subplot(121) exp_x = stats.expon.pdf(x, scale=3) exp_y = stats.expon.pdf(x, scale=10) M = np.dot(exp_x[:, None], exp_y[None, :]) CS = plt.contour(X, Y, M) # 等高線をプロットするようだ。 # ここについても、座標の取り方はax.plot_surface(X, Y, M, cmap=plt.cm.jet, vmax=1, vmin=-0.15) # と同じように理解できる。 im = plt.imshow(M, interpolation='none', origin='lower', cmap=jet, extent=(0, 5, 0, 5)) plt.title("Overhead view of landscape formed by " "$Exp(3), Exp(10)$ priors") ax = fig.add_subplot(122, projection='3d') ax.plot_surface(X, Y, M, cmap=jet) ax.view_init(azim=390) ax.set_xlabel('Value of $p_1$') ax.set_ylabel('Value of $p_2$') ax.set_zlabel('Density') plt.title("Alternate view of landscape\nformed by " "Exp(3), Exp(10) priors")
Output:
Text(0.5, 0.92, 'Alternate view of landscape\nformed by Exp(3), Exp(10) priors')

def plot_landscape_warped_by_n_data_obs_Exp_pri_dist(N=1): lambda_1_true = 1 lambda_2_true = 3 plt.rcParams['font.size'] = 17 data = np.concatenate([ stats.poisson.rvs(lambda_1_true, size=(N, 1)), stats.poisson.rvs(lambda_2_true, size=(N, 1)) ], axis=1) print(f"observed (2-dimensional, sample size = {N}):", data) x = y = np.linspace(.01, 5, 100) X, Y = np.meshgrid(x, y) likelihood_x = np.array([stats.poisson.pmf(data[:, 0], _x) for _x in x]).prod(axis=1) likelihood_y = np.array([stats.poisson.pmf(data[:, 1], _y) for _y in y]).prod(axis=1) L = np.dot(likelihood_x[:, None], likelihood_y[None, :]) figsize(25, 24) plt.subplot(221) uni_x = stats.uniform.pdf(x, loc=0, scale=5) uni_y = stats.uniform.pdf(y, loc=0, scale=5) M = np.dot(uni_x[:, None], uni_y[None, :]) plt.imshow(M, interpolation='none', origin='lower', cmap=jet, vmax=1, vmin=-.15, extent=(0, 5, 0, 5)) plt.scatter(lambda_2_true, lambda_1_true, c='k', s=50, edgecolor="none") plt.xlim(0, 5) plt.ylim(0, 5) plt.title("Landscape formed by " "Uniform priors on $p_1, p_2$") plt.xlabel("Value of $p_1$") plt.ylabel("Value of $p_2$") plt.subplot(223) plt.contour(X, Y, M * L) plt.imshow(M * L, interpolation='none', origin='lower', cmap=jet, extent=(0, 5, 0, 5)) plt.scatter(lambda_2_true, lambda_1_true, c='k', s=50, edgecolor='none') plt.xlim(0, 5) plt.ylim(0, 5) plt.title(f"Lnadscape warped by {N} data observation;\n" "Uniform priors on $p_1, p_2$") plt.xlabel("Value of $p_1$") plt.ylabel("Value of $p_2$") plt.subplot(222) exp_x = stats.expon.pdf(x, loc=0, scale=3) exp_y = stats.expon.pdf(y, loc=0, scale=10) M = np.dot(exp_x[:, None], exp_y[None, :]) plt.contour(x, y, M) plt.imshow(M, interpolation='none', origin='lower', cmap=jet, extent=(0, 5, 0, 5)) plt.scatter(lambda_2_true, lambda_1_true, c="k", s=50, edgecolor="none") plt.xlim(0, 5) plt.ylim(0, 5) plt.title("Landscape formed by " "Exponential priors on $p_1, p_2$") plt.xlabel("Value of $p_1$") plt.ylabel("Value of $p_2$") plt.subplot(224) plt.contour(x, y, M * L) plt.imshow(M * L, interpolation='none', origin='lower', cmap=jet, extent=(0, 5, 0, 5)) plt.scatter(lambda_2_true, lambda_1_true, c="k", s=50, edgecolor="none") plt.xlim(0, 5) plt.ylim(0, 5) plt.xlabel('Value of $p_1$') plt.ylabel('Value of $p_2$') plt.title(f"Landscape warped by {N} data observation;\n" "Exponential priors on $p_1, p_2$") plt.xlabel("Value of $p_1$") plt.ylabel("Value of $p_2$")
plot_landscape_warped_by_n_data_obs_Exp_pri_dist()
Output:
observed (2-dimensional, sample size = 1): [[2 1]]

plot_landscape_warped_by_n_data_obs_Exp_pri_dist(N=2)
Output:
observed (2-dimensional, sample size = 2): [[2 2]
[1 4]]

メモ
関数plot_landscape_warped_by_n_data_obs_Exp_pri_distの以下の部分でどのようなarrayが生成されているのかをメモしておきます。
data = np.concatenate([
stats.poisson.rvs(lambda_1_true, size=(N, 1)),
stats.poisson.rvs(lambda_2_true, size=(N, 1))
], axis=1)
print(f"observed (2-dimensional, sample size = {N}):", data)
x = y = np.linspace(.01, 5, 100)
X, Y = np.meshgrid(x, y)
likelihood_x = np.array([stats.poisson.pmf(data[:, 0], _x) for _x in x]).prod(axis=1)
likelihood_y = np.array([stats.poisson.pmf(data[:, 1], _y) for _y in y]).prod(axis=1)
N=2 lambda_1_true = 1 lambda_2_true = 3 data = np.concatenate([ stats.poisson.rvs(lambda_1_true, size=(N, 1)), stats.poisson.rvs(lambda_2_true, size=(N, 1)) ], axis=1) print('data:') pprint.pprint(data) x = y = np.linspace(.01, 5, 100) print('\ndata[:, 0]:') pprint.pprint(data[:, 0]) likelihood_x = np.array([stats.poisson.pmf(data[:, 0], _x) for _x in x]).prod(axis=1) print('\nnp.array([stats.poisson.pmf(data[:, 0], _x) for _x in x]):') pprint.pprint(np.array([stats.poisson.pmf(data[:, 0], _x) for _x in x])) print('\nnp.array([stats.poisson.pmf(data[:, 0], _x) for _x in x]).prod(axis=1):') pprint.pprint(np.array([stats.poisson.pmf(data[:, 0], _x) for _x in x]).prod(axis=1))
Output:(クリックすると展開されます)
data:
array([[ 2, 10],
[ 0, 4]])
data[:, 0]:
array([2, 0])
np.array([stats.poisson.pmf(data[:, 0], _x) for _x in x]):
array([[ 4.95024917e-05, 9.90049834e-01],
[ 1.71738965e-03, 9.41384100e-01],
[ 5.49527628e-03, 8.95110521e-01],
[ 1.10599167e-02, 8.51111513e-01],
[ 1.81202397e-02, 8.09275269e-01],
[ 2.64146968e-02, 7.69495478e-01],
[ 3.57088061e-02, 7.31671056e-01],
[ 4.57928792e-02, 6.95705888e-01],
[ 5.64799179e-02, 6.61508581e-01],
[ 6.76036698e-02, 6.28992237e-01],
[ 7.90168306e-02, 5.98074229e-01],
[ 9.05893832e-02, 5.68675990e-01],
[ 1.02207064e-01, 5.40722817e-01],
[ 1.13769950e-01, 5.14143677e-01],
[ 1.25191149e-01, 4.88871030e-01],
[ 1.36395603e-01, 4.64840657e-01],
[ 1.47318973e-01, 4.41991492e-01],
[ 1.57906624e-01, 4.20265474e-01],
[ 1.68112684e-01, 3.99607395e-01],
[ 1.77899183e-01, 3.79964760e-01],
[ 1.87235256e-01, 3.61287656e-01],
[ 1.96096421e-01, 3.43528621e-01],
[ 2.04463907e-01, 3.26642529e-01],
[ 2.12324047e-01, 3.10586470e-01],
[ 2.19667714e-01, 2.95319644e-01],
[ 2.26489810e-01, 2.80803256e-01],
[ 2.32788800e-01, 2.67000419e-01],
[ 2.38566283e-01, 2.53876058e-01],
[ 2.43826605e-01, 2.41396823e-01],
[ 2.48576502e-01, 2.29531002e-01],
[ 2.52824780e-01, 2.18248444e-01],
[ 2.56582021e-01, 2.07520479e-01],
[ 2.59860318e-01, 1.97319844e-01],
[ 2.62673037e-01, 1.87620620e-01],
[ 2.65034598e-01, 1.78398160e-01],
[ 2.66960278e-01, 1.69629028e-01],
[ 2.68466040e-01, 1.61290942e-01],
[ 2.69568370e-01, 1.53362712e-01],
[ 2.70284140e-01, 1.45824194e-01],
[ 2.70630477e-01, 1.38656230e-01],
[ 2.70624657e-01, 1.31840606e-01],
[ 2.70283997e-01, 1.25360003e-01],
[ 2.69625775e-01, 1.19197953e-01],
[ 2.68667148e-01, 1.13338798e-01],
[ 2.67425086e-01, 1.07767648e-01],
[ 2.65916310e-01, 1.02470347e-01],
[ 2.64157245e-01, 9.74334344e-02],
[ 2.62163976e-01, 9.26441102e-02],
[ 2.59952207e-01, 8.80902044e-02],
[ 2.57537235e-01, 8.37601451e-02],
[ 2.54933921e-01, 7.96429292e-02],
[ 2.52156669e-01, 7.57280943e-02],
[ 2.49219412e-01, 7.20056925e-02],
[ 2.46135598e-01, 6.84662647e-02],
[ 2.42918183e-01, 6.51008169e-02],
[ 2.39579622e-01, 6.19007971e-02],
[ 2.36131869e-01, 5.88580738e-02],
[ 2.32586373e-01, 5.59649150e-02],
[ 2.28954087e-01, 5.32139689e-02],
[ 2.25245463e-01, 5.05982452e-02],
[ 2.21470464e-01, 4.81110969e-02],
[ 2.17638567e-01, 4.57462039e-02],
[ 2.13758775e-01, 4.34975569e-02],
[ 2.09839623e-01, 4.13594418e-02],
[ 2.05889189e-01, 3.93264253e-02],
[ 2.01915108e-01, 3.73933415e-02],
[ 1.97924578e-01, 3.55552780e-02],
[ 1.93924380e-01, 3.38075643e-02],
[ 1.89920884e-01, 3.21457591e-02],
[ 1.85920064e-01, 3.05656397e-02],
[ 1.81927514e-01, 2.90631907e-02],
[ 1.77948459e-01, 2.76345945e-02],
[ 1.73987767e-01, 2.62762206e-02],
[ 1.70049965e-01, 2.49846173e-02],
[ 1.66139254e-01, 2.37565027e-02],
[ 1.62259517e-01, 2.25887557e-02],
[ 1.58414338e-01, 2.14784092e-02],
[ 1.54607011e-01, 2.04226416e-02],
[ 1.50840556e-01, 1.94187701e-02],
[ 1.47117728e-01, 1.84642437e-02],
[ 1.43441034e-01, 1.75566369e-02],
[ 1.39812739e-01, 1.66936433e-02],
[ 1.36234886e-01, 1.58730701e-02],
[ 1.32709300e-01, 1.50928319e-02],
[ 1.29237602e-01, 1.43509463e-02],
[ 1.25821221e-01, 1.36455279e-02],
[ 1.22461403e-01, 1.29747843e-02],
[ 1.19159221e-01, 1.23370109e-02],
[ 1.15915588e-01, 1.17305872e-02],
[ 1.12731260e-01, 1.11539722e-02],
[ 1.09606852e-01, 1.06057005e-02],
[ 1.06542842e-01, 1.00843791e-02],
[ 1.03539583e-01, 9.58868312e-03],
[ 1.00597308e-01, 9.11735301e-03],
[ 9.77161399e-02, 8.66919104e-03],
[ 9.48960974e-02, 8.24305840e-03],
[ 9.21371035e-02, 7.83787224e-03],
[ 8.94389915e-02, 7.45260294e-03],
[ 8.68015116e-02, 7.08627149e-03],
[ 8.42243375e-02, 6.73794700e-03]])
np.array([stats.poisson.pmf(data[:, 0], _x) for _x in x]).prod(axis=1):
array([ 4.90099337e-05, 1.61672331e-03, 4.91887962e-03,
9.41322241e-03, 1.46642619e-02, 2.03259898e-02,
2.61270999e-02, 3.18583757e-02, 3.73619503e-02,
4.25221835e-02, 4.72579300e-02, 5.15160072e-02,
5.52656917e-02, 5.84941004e-02, 6.12023262e-02,
6.34022216e-02, 6.51137326e-02, 6.63627021e-02,
6.71790718e-02, 6.75954202e-02, 6.76457866e-02,
6.73647330e-02, 6.67866079e-02, 6.59449765e-02,
6.48721911e-02, 6.35990761e-02, 6.21547071e-02,
6.05662675e-02, 5.88589677e-02, 5.70560137e-02,
5.51786149e-02, 5.32460237e-02, 5.12755975e-02,
4.92828782e-02, 4.72816845e-02, 4.52842125e-02,
4.33011403e-02, 4.13417363e-02, 3.94139667e-02,
3.75246016e-02, 3.56793187e-02, 3.38828027e-02,
3.21388405e-02, 3.04504115e-02, 2.88197725e-02,
2.72485366e-02, 2.57377476e-02, 2.42879483e-02,
2.28992431e-02, 2.15713562e-02, 2.03036842e-02,
1.90953440e-02, 1.79452163e-02, 1.68519850e-02,
1.58141722e-02, 1.48301696e-02, 1.38982669e-02,
1.30166766e-02, 1.21835557e-02, 1.13970252e-02,
1.06551869e-02, 9.95613829e-03, 9.29798450e-03,
8.67884968e-03, 8.09688583e-03, 7.55028057e-03,
7.03726341e-03, 6.55611095e-03, 6.10515098e-03,
5.68276569e-03, 5.28739405e-03, 4.91753350e-03,
4.57174093e-03, 4.24863331e-03, 3.94688762e-03,
3.66524060e-03, 3.40248798e-03, 3.15748358e-03,
2.92913807e-03, 2.71641758e-03, 2.51834214e-03,
2.33398400e-03, 2.16246589e-03, 2.00295916e-03,
1.85468189e-03, 1.71689698e-03, 1.58891028e-03,
1.47006862e-03, 1.35975791e-03, 1.25740133e-03,
1.16245744e-03, 1.07441841e-03, 9.92808253e-04,
9.17181170e-04, 8.47119885e-04, 7.82234073e-04,
7.22158846e-04, 6.66553291e-04, 6.15099077e-04,
5.67499122e-04])
3.1.4 例題:混合モデルの教師無しクラスタリング
makedirs("data", exist_ok=True) urlretrieve("https://git.io/vXt6b", "data/mixture_data.csv")
Output:
('data/mixture_data.csv', <http.client.HTTPMessage at 0x1606930ab38>)
figsize(25, 8) plt.rcParams['font.size'] = 17 data = np.loadtxt("data/mixture_data.csv", delimiter=",") plt.hist(data, bins=20, color="k", histtype="stepfilled", alpha=0.8) plt.title("Histogram of the dataset") plt.ylim([0, None]) plt.xlabel('Value') plt.ylabel('Count') print(data[:10], "...")
Output:
[ 115.85679142 152.26153716 178.87449059 162.93500815 107.02820697
105.19141146 118.38288501 125.3769803 102.88054011 206.71326136] ...

p = pm.Uniform("p", 0., 1.) assignment = pm.Categorical("assignment", [p, 1-p], size=data.shape[0]) taus = 1.0 / pm.Uniform("stds", lower=0, upper=100, size=2) ** 2 # テキストを読んだ感じ、ここはupper=100なんじゃないんかな? centers = pm.Normal("centers", [120, 190], [0.01, 0.01], size=2) @pm.deterministic def center_i(assignment=assignment, centers=centers): return centers[assignment] @pm.deterministic def tau_i(assignment=assignment, taus=taus): return taus[assignment]
どうやら、centers[assignment]のように書くと、assignmentと同じサイズのサンプルを返してくれるらしい。しかも、各平均はちゃんとassignmentでサンプルされたクラスタからサンプルされるらしい。すごい・・・
print(f"Random assignment: {assignment.value[:8]}") print(f"Assigned center: {center_i.value[:8]}") print(f"Assigned precision: {tau_i.value[:8]}")
Output:
Random assignment: [0 1 1 0 0 0 1 1]
Assigned center: [ 128.32964549 191.13553451 191.13553451 128.32964549 128.32964549
128.32964549 191.13553451 191.13553451]
Assigned precision: [ 0.0001068 0.00012591 0.00012591 0.0001068 0.0001068 0.0001068
0.00012591 0.00012591]
observations = pm.Normal("obs", center_i, tau_i, value=data, observed=True) # これ、どこで使うんだ? model = pm.Model([p, assignment, taus, centers])
mcmc = pm.MCMC(model)
mcmc.sample(50000)
Output:
[-----------------100%-----------------] 50000 of 50000 complete in 20.9 sec
figsize(25, 18) plt.rcParams['font.size'] = 17 line_width = 1 colors = ["#348ABD", "#A60628"] center_trace = mcmc.trace("centers")[:] if center_trace[-1, 0] < center_trace[-1, 1]: colors = ["#A60628", "#348ABD"] plt.subplot(311) plt.plot(center_trace[:, 0], label="trace of center 0", c=colors[0], lw=line_width) plt.plot(center_trace[:, 1], label="trace of center 1", c=colors[1], lw=line_width) plt.title("Traces of unknown parameters") leg = plt.legend(loc="upper right") leg.get_frame().set_alpha(0.7) plt.subplot(312) std_trace = mcmc.trace("stds")[:] plt.plot(std_trace[:, 0], label="trace of standard deviation " "of cluster 0", c=colors[0], lw=line_width) plt.plot(std_trace[:, 1], label="trace of standard deviation " "of cluster 1", c=colors[1], lw=line_width) plt.legend(loc="upper right") plt.subplot(313) p_trace = mcmc.trace("p")[:] plt.plot(p_trace, label="$p$: frequency of assignment to cluster 0", color="#467821", lw=line_width) plt.xlabel("Steps") plt.ylabel("Value") plt.ylim(0, 1) plt.legend()
Output:
<matplotlib.legend.Legend at 0x16020cd9828>

mcmc.sample(100000)
Output:
[-----------------100%-----------------] 100000 of 100000 complete in 41.7 sec
figsize(25, 8) plt.rcParams['font.size'] = 17 center_trace = mcmc.trace("centers", chain=1)[:] prev_center_trace = mcmc.trace("centers", chain=0)[:] x = np.arange(50000) plt.plot(x, prev_center_trace[:, 0], label="previous trace of center 0", lw=line_width, alpha=0.4, c=colors[0]) plt.plot(x, prev_center_trace[:, 1], label="previous trace of center 1", lw=line_width, alpha=0.4, c=colors[1]) x = np.arange(50000, 150000) plt.plot(x, center_trace[:, 0], label="new trace of center 0", lw=line_width, c=colors[0]) plt.plot(x, center_trace[:, 1], label="new trace of center 1", lw=line_width, c=colors[1]) plt.title("Trace of unknown center parameters " "after sampling 1000,000 more times") leg = plt.legend(loc="upper right") leg.get_frame().set_alpha(0.8) plt.ylabel("Value") plt.xlabel("Steps")
Output:
Text(0.5, 0, 'Steps')

figsize(25, 9) plt.rcParams['font.size'] = 17 plt.cmap = mpl.colors.ListedColormap(colors) plt.imshow(mcmc.trace("assignment")[::400, np.argsort(data)], # np.argsort(data)を引数に持たせることで、各サンプルを観測データが小さい順に並べ替えてくれる。 cmap=plt.cmap, aspect=.4, alpha=.9) plt.xticks(np.arange(0, data.shape[0], 40), [f"{round(s, 2)}" for s in np.sort(data)[::40]]) plt.ylabel("Posterior sample") plt.xlabel("Value of $i$th data point") plt.title("Posterior labels fo data points")
Output:
Text(0.5, 1.0, 'Posterior labels fo data points')

cmap = mpl.colors.LinearSegmentedColormap.from_list("BMH", colors) assign_trace = mcmc.trace("assignment")[:] plt.scatter(data, 1 - assign_trace.mean(axis=0), cmap=cmap, c=assign_trace.mean(axis=0), s=50) plt.ylim(-0.05, 1.05) plt.xlim(35, 300) plt.title("Probability of data point belonging to cluster 0") plt.ylabel("Probability") plt.xlabel("Value of data point")
Output:
Text(0.5, 0, 'Value of data point')

norm = stats.norm x = np.linspace(20, 300, 500) posterior_center_means = center_trace.mean(axis=0) posterior_std_means = std_trace.mean(axis=0) posterior_p_mean = mcmc.trace("p")[:].mean() plt.hist(data, bins=20, histtype="step", density=True, color="k", lw=2, label="histogram of data") y = posterior_p_mean * norm.pdf(x, loc=posterior_center_means[0], scale=posterior_std_means[0]) plt.plot(x, y, lw=3, color=colors[0], label="cluster 0" "(using posterior-mean parameters)") plt.fill_between(x, y, color=colors[0], alpha=0.3) y = (1 - posterior_p_mean) * norm.pdf(x, loc=posterior_center_means[1], scale=posterior_std_means[1]) plt.plot(x, y, lw=3, color=colors[1], label="cluster 1" "(using posterior-mean parameters)") plt.fill_between(x, y, color=colors[1], alpha=0.3) plt.legend(loc="upper left") plt.title("Visualizing clusters using posterior-mean parameters")
Output:
Text(0.5, 1.0, 'Visualizing clusters using posterior-mean parameters')

3.1.5 事後サンプルを混ぜないで
p_trace=mcmc.trace("p")[:] std_trace = mcmc.trace("stds")[:]
def prob_of_belong_cluster(x, p_trace=p_trace, std_trace=std_trace): norm_pdf = stats.norm.pdf v = p_trace * norm_pdf(x, loc=center_trace[:, 0], scale=std_trace[:, 0]) < \ (1 - p_trace) * norm_pdf(x, loc=center_trace[:, 1], scale=std_trace[:, 1]) # テキストは不等号が逆なんじゃないか? return v.mean()
X = np.linspace(20, 300, 500) Y = np.array([prob_of_belong_cluster(x=x) for x in X])
各xの値について、クラスタ1に属する確率をプロットしてみた。
figsize(25, 9) fig, ax1 = plt.subplots() ax1.hist(data, bins=20, histtype="step", density=True, color="k", lw=2, label="histogram of data") ax2 = ax1.twinx() # 2つのプロットを関連付ける ax2.plot(X, Y, label="Probability of belonging to cluster 1") plt.ylim(0, 1.01) plt.legend(loc="best")
Output:
<matplotlib.legend.Legend at 0x160037e7b00>

3.1.6 MAPを使って収束を改善
pm.MAPを使用してみました。だけど、使い方が間違えてるのか、バーンインが短くなっているかどうかはぱっと見わかりませんね・・・
pの初期値なんてほぼ0になってるし・・・
p = pm.Uniform("p", 0., 1.) assignment = pm.Categorical("assignment", [p, 1-p], size=data.shape[0]) taus = 1.0 / pm.Uniform("stds", lower=0, upper=100, size=2) ** 2 # テキストを読んだ感じ、ここはupper=100なんじゃないんかな? centers = pm.Normal("centers", [120, 190], [0.01, 0.01], size=2) @pm.deterministic def center_i(assignment=assignment, centers=centers): return centers[assignment] @pm.deterministic def tau_i(assignment=assignment, taus=taus): return taus[assignment] observations = pm.Normal("obs", center_i, tau_i, value=data, observed=True) # これ、どこで使うんだ? model = pm.Model([p, assignment, taus, centers]) map_ = pm.MAP(model) map_.fit() mcmc = pm.MCMC(model) mcmc.sample(50000) figsize(25, 18) plt.rcParams['font.size'] = 17 line_width = 1 colors = ["#348ABD", "#A60628"] center_trace = mcmc.trace("centers")[:] if center_trace[-1, 0] < center_trace[-1, 1]: colors = ["#A60628", "#348ABD"] plt.subplot(311) plt.plot(center_trace[:, 0], label="trace of center 0", c=colors[0], lw=line_width) plt.plot(center_trace[:, 1], label="trace of center 1", c=colors[1], lw=line_width) plt.title("Traces of unknown parameters") leg = plt.legend(loc="upper right") leg.get_frame().set_alpha(0.7) plt.subplot(312) std_trace = mcmc.trace("stds")[:] plt.plot(std_trace[:, 0], label="trace of standard deviation " "of cluster 0", c=colors[0], lw=line_width) plt.plot(std_trace[:, 1], label="trace of standard deviation " "of cluster 1", c=colors[1], lw=line_width) plt.legend(loc="upper right") plt.subplot(313) p_trace = mcmc.trace("p")[:] plt.plot(p_trace, label="$p$: frequency of assignment to cluster 0", color="#467821", lw=line_width) plt.xlabel("Steps") plt.ylabel("Value") plt.ylim(0, 1) plt.legend()
Output:
Warning: Stochastic assignment's value is neither numerical nor array with floating-point dtype. Recommend fitting method fmin (default).
[-----------------100%-----------------] 50000 of 50000 complete in 19.7 sec
<matplotlib.legend.Legend at 0x160049da240>

今回の写経は以上です。 ここまで読んでいただきありがとうございました。
Pythonで体験するベイズ推論 PyMCによるMCMC入門 第2章 (2.2.6から2章の終わりまで) 写経
目次
始めに
この記事では、Pythonで体験するベイズ推論 PyMCによるMCMC入門
の第2章の2.2.4までを写経した記録をまとめます。前回はこちらPythonで体験するベイズ推論 PyMCによるMCMC入門 第2章 (2.2.4まで) 写経。 実行はJupyter Labにて実行しています。
写経は以下のようにまとめました。
写経ではありますが、コードを実行しやすいように関数を作成し、コードの再実行がやりやすいようにした部分があります。
また、より良いと思われるコードを考えた場合は書き換えてコメントを添えるようにし、変更点をなるべく明示するようにしてあります。
個人的に気になった点のメモを残すようにしてあります。同じような疑問を持った方の助けになれば幸いです。
以前書いたコードと同じようなコード(例えばグラフの描写等)は効率化のために飛ばしているところもあります。
Pythonで体験するベイズ推論 PyMCによるMCMC入門を読んでいている際のちょっとした正誤表代わりになればと思います。
この記事で使用する主なモジュール、設定
この記事では、主に以下のモジュールや設定を使用しています。
import scipy.stats as stats from IPython.core.pylabtools import figsize from matplotlib import pyplot as plt %matplotlib inline import numpy as np plt.rcParams['font.size'] = 17 import pymc as pm from scipy.stats.mstats import mquantiles from urllib.request import urlretrieve from os import makedirs
2 PyMCについてもう少し
2.2 モデリングのアプローチ
2.2.6 二項分布
def plot_bin_dist_pmf(N=10, means=[.4, .9]): figsize(25, 8) binomial = stats.binom for p in means: _x = np.arange(N + 1) plt.bar(_x, binomial.pmf(_x, N, p), alpha=0.6, linewidth=3, label=f"$N$: {N}, $p$: {p}") plt.legend(loc="upper left") plt.xlim(-.5, N + 0.5) plt.xlabel("$k$") plt.ylabel("$P(X = k)$") plt.title("Probability mass distributions of " "binomial random variables")
plot_bin_dist_pmf()
Output:

以下の図は、二項分布が正規分布に収束するのが実感できますね。
plot_bin_dist_pmf(N=1000, means=[.5, .8])
Output:

2.2.7 例題:カンニングをした学生の割合
N = 100 p = pm.Uniform("freq_cheating", 0, 1) true_answers = pm.Bernoulli("truths", p, size=N) first_coin_flips = pm.Bernoulli("first_flips", 0.5, size=N) print(first_coin_flips.value)
Output:
[False True True False False True False True True False False False
True True False True False False True False True True False False
True False False False True False True True False True True True
False True True True True False False False False False False False
False True True True False True False False False False False False
True False False True True False True False True False True False
False True True True False True True True True True False True
False True True True False True False True True True True False
True True False True]
second_coin_flips = pm.Bernoulli("second_flips", 0.5, size=N) @pm.deterministic def observed_proportion(t_a=true_answers, fc=first_coin_flips, sc=second_coin_flips): observed = fc * t_a + (1 - fc) * sc return observed.sum()/float(N) print(observed_proportion.value)
Output:
0.41
X = 35 observations = pm.Binomial("obs", N, observed_proportion, observed=True, value=X) model = pm.Model([p, true_answers, first_coin_flips, second_coin_flips, observed_proportion, observations]) mcmc = pm.MCMC(model) mcmc.sample(40000, 15000)
Output:
[-----------------100%-----------------] 40000 of 40000 complete in 13.5 sec
figsize(25, 6) p_trace = mcmc.trace("freq_cheating")[:] plt.hist(p_trace, histtype="stepfilled", density=True, alpha=0.85, bins=30, color="#348ABD", label="posterior distribution") plt.vlines([.05, .35], [0, 0], [5, 5], alpha=0.3) plt.xlim(0, 1) plt.xlabel("Value of $p$") plt.ylabel("Density") plt.title("Posterior distribution of parameter $p$") plt.legend()
Output:
<matplotlib.legend.Legend at 0x1fae1706320>

2.2.8 もう一つのPyMCモデル
p = pm.Uniform("freq_cheating", 0, 1) @pm.deterministic def p_skewed(p=p): return 0.5 * p + 0.25 yes_responses = pm.Binomial("number_cheaters", 100, p_skewed, value=35, observed=True) model = pm.Model([yes_responses, p_skewed, p]) mcmc = pm.MCMC(model) mcmc.sample(25000, 2500)
Output:
[-----------------100%-----------------] 25000 of 25000 complete in 1.2 sec
figsize(25, 6) p_trace = mcmc.trace("freq_cheating")[:] plt.hist(p_trace, histtype="stepfilled", density=True, alpha=0.85, bins=30, color="#348ABD", label="posterior distribution") plt.vlines([.05, .35], [0, 0], [5, 5], alpha=0.3) plt.xlim(0, 1) plt.xlabel("Value of $p$") plt.ylabel("Density") plt.title("Posterior distribution of parameter $p$" "from alternate model") plt.legend()
Output:
<matplotlib.legend.Legend at 0x1fadfc56eb8>

2.2.9 PyMCの使い方をもう少し
上級者向け:Lambdaクラスを使ってお手軽にdeterministic変数を扱う
beta = pm.Normal("coefficients", 0, 1, size=(N, 1)) x = np.random.randn(N, 1)
メモ
np.random.randnは標準正規分布からのサンプルを返す関数のようです。
引数の整数は返されるarrayオブジェクトの次元を指定するそうです。
np.random.randn??(クリックすると展開されます)
Docstring:
randn(d0, d1, ..., dn)
Return a sample (or samples) from the "standard normal" distribution.
If positive, int_like or int-convertible arguments are provided,
`randn` generates an array of shape ``(d0, d1, ..., dn)``, filled
with random floats sampled from a univariate "normal" (Gaussian)
distribution of mean 0 and variance 1 (if any of the :math:`d_i` are
floats, they are first converted to integers by truncation). A single
float randomly sampled from the distribution is returned if no
argument is provided.
This is a convenience function. If you want an interface that takes a
tuple as the first argument, use `numpy.random.standard_normal` instead.
Parameters
----------
d0, d1, ..., dn : int, optional
The dimensions of the returned array, should be all positive.
If no argument is given a single Python float is returned.
Returns
-------
Z : ndarray or float
A ``(d0, d1, ..., dn)``-shaped array of floating-point samples from
the standard normal distribution, or a single such float if
no parameters were supplied.
See Also
--------
random.standard_normal : Similar, but takes a tuple as its argument.
Notes
-----
For random samples from :math:`N(\mu, \sigma^2)`, use:
``sigma * np.random.randn(...) + mu``
Examples
--------
>>> np.random.randn()
2.1923875335537315 #random
Two-by-four array of samples from N(3, 6.25):
>>> 2.5 * np.random.randn(2, 4) + 3
array([[-4.49401501, 4.00950034, -1.81814867, 7.29718677], #random
[ 0.39924804, 4.68456316, 4.99394529, 4.84057254]]) #random
Type: builtin_function_or_method
linear_combination = pm.Lambda("comb", lambda x=x, beta=beta: np.dot(x.T, beta))
xとbetaの内積を計算したdeterministic変数が作成されます。
linear_combination.value
Output:
array([[ 3.54607155]])
メモ
x.Tはxの転置行列(ベクトル?)を返します。
x.T
Output:
array([[-0.69877594, -0.30683497, -1.04543408, 0.92360732, -0.84359431,
-0.8104658 , 1.033511 , 1.06662984, -0.87370605, -1.61567741,
0.277752 , 0.06679146, 1.05671416, -0.04508463, -0.87050541,
-0.29798197, 1.41862678, -0.18613036, -1.14649344, -3.38937616,
-0.34949099, 0.26877773, 1.61307278, 0.11562269, -0.18933638,
0.0502833 , -0.46785645, 1.40923847, -0.50753077, 0.18046069,
1.46627307, 1.14276522, -0.31624792, -0.13193391, 1.00854897,
0.53726617, 0.75892404, -0.49679437, -0.49542055, -0.29550763,
0.38086154, -0.22335623, 0.42129412, -0.27474526, -1.06314766,
-0.25977146, -0.12190971, 1.61424116, -0.7144213 , -0.11599007,
1.78002587, -0.34226 , -0.02305097, -0.22823814, 0.26100848,
-0.39558157, -1.78600444, 0.38610165, -1.14085662, -0.9320066 ,
-0.55027475, -0.1132584 , -0.26211372, -0.05095378, 1.59816836,
0.61105632, -1.26027571, 1.28556484, -1.61263487, -0.62419771,
-0.01766886, 2.28490072, -0.82740689, -0.03267927, -0.45830663,
1.51543338, -0.39441128, -0.13035409, -0.4335249 , -0.70002868,
1.36377425, -1.83376874, -0.44147943, 0.00362854, -1.01908356,
2.54583705, -0.12141319, 0.27333106, 2.22923404, 0.1929837 ,
-2.02996548, -0.29450739, 0.22175473, 2.27184055, 0.39794602,
0.53896599, -0.46472929, -1.58689283, 1.6319995 , 0.86209208]])
np.dotはベクトル同士の内積や行列の積を計算します。以下では5行1列の行列と1行5列の行列を掛け合わせ、5行5列の行列がアウトプットされています。
np.dot(x[:5], x[:5].T)
Output:
array([[ 0.48828781, 0.2144089 , 0.73052418, -0.64539457, 0.58948341],
[ 0.2144089 , 0.0941477 , 0.32077574, -0.28339503, 0.25884424],
[ 0.73052418, 0.32077574, 1.09293241, -0.96557056, 0.88192224],
[-0.64539457, -0.28339503, -0.96557056, 0.85305048, -0.77914988],
[ 0.58948341, 0.25884424, 0.88192224, -0.77914988, 0.71165136]])
np.dotの説明はこんな感じ。
np.dot??(クリックすると展開されます)
Docstring:
dot(a, b, out=None)
Dot product of two arrays.
For 2-D arrays it is equivalent to matrix multiplication, and for 1-D
arrays to inner product of vectors (without complex conjugation). For
N dimensions it is a sum product over the last axis of `a` and
the second-to-last of `b`::
dot(a, b)[i,j,k,m] = sum(a[i,j,:] * b[k,:,m])
Parameters
----------
a : array_like
First argument.
b : array_like
Second argument.
out : ndarray, optional
Output argument. This must have the exact kind that would be returned
if it was not used. In particular, it must have the right type, must be
C-contiguous, and its dtype must be the dtype that would be returned
for `dot(a,b)`. This is a performance feature. Therefore, if these
conditions are not met, an exception is raised, instead of attempting
to be flexible.
Returns
-------
output : ndarray
Returns the dot product of `a` and `b`. If `a` and `b` are both
scalars or both 1-D arrays then a scalar is returned; otherwise
an array is returned.
If `out` is given, then it is returned.
Raises
------
ValueError
If the last dimension of `a` is not the same size as
the second-to-last dimension of `b`.
See Also
--------
vdot : Complex-conjugating dot product.
tensordot : Sum products over arbitrary axes.
einsum : Einstein summation convention.
matmul : '@' operator as method with out parameter.
Examples
--------
>>> np.dot(3, 4)
12
Neither argument is complex-conjugated:
>>> np.dot([2j, 3j], [2j, 3j])
(-13+0j)
For 2-D arrays it is the matrix product:
>>> a = [[1, 0], [0, 1]]
>>> b = [[4, 1], [2, 2]]
>>> np.dot(a, b)
array([[4, 1],
[2, 2]])
>>> a = np.arange(3*4*5*6).reshape((3,4,5,6))
>>> b = np.arange(3*4*5*6)[::-1].reshape((5,4,6,3))
>>> np.dot(a, b)[2,3,2,1,2,2]
499128
>>> sum(a[2,3,2,:] * b[1,2,:,2])
499128
Type: builtin_function_or_method
上級者向け:PyMC変数のarray
N = 10 x = np.empty(N, dtype=object) for i in range(0, N): x[i] = pm.Exponential(f"x_{i}", (i + 1)**2) x
Output:
array([ <pymc.distributions.new_dist_class.<locals>.new_class 'x_0' at 0x000001FADFA052B0>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_1' at 0x000001FADFA05240>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_2' at 0x000001FADFA051D0>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_3' at 0x000001FADFA05160>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_4' at 0x000001FADFA050F0>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_5' at 0x000001FADFA05080>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_6' at 0x000001FADFA05FD0>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_7' at 0x000001FADFA05A20>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_8' at 0x000001FADFA05EF0>,
<pymc.distributions.new_dist_class.<locals>.new_class 'x_9' at 0x000001FADFA05D30>], dtype=object)
xは各成分がPyMC変数となっているarrayオブジェクトになっています。
print(x[0].value) x[0].random() print(x[0].value)
Output:
1.347759419712658
0.6845365343616834
メモ
ちなみに、dtype=objectを指定しないと、指数分布からサンプリングされた値がxに保存されるようです。
N = 10 x = np.empty(N) for i in range(0, N): x[i] = pm.Exponential(f"x_{i}", (i + 1)**2) x
Output:
array([ 0.76906025, 0.14474833, 0.11372349, 0.04600305, 0.038043 ,
0.01263401, 0.02176758, 0.00217072, 0.00591296, 0.0207582 ])
np.emptyの説明はこんな感じ。
np.empty??(クリックすると展開されます)
Docstring:
empty(shape, dtype=float, order='C')
Return a new array of given shape and type, without initializing entries.
Parameters
----------
shape : int or tuple of int
Shape of the empty array
dtype : data-type, optional
Desired output data-type.
order : {'C', 'F'}, optional
Whether to store multi-dimensional data in row-major
(C-style) or column-major (Fortran-style) order in
memory.
Returns
-------
out : ndarray
Array of uninitialized (arbitrary) data of the given shape, dtype, and
order. Object arrays will be initialized to None.
See Also
--------
empty_like, zeros, ones
Notes
-----
`empty`, unlike `zeros`, does not set the array values to zero,
and may therefore be marginally faster. On the other hand, it requires
the user to manually set all the values in the array, and should be
used with caution.
Examples
--------
>>> np.empty([2, 2])
array([[ -9.74499359e+001, 6.69583040e-309],
[ 2.13182611e-314, 3.06959433e-309]]) #random
>>> np.empty([2, 2], dtype=int)
array([[-1073741821, -1067949133],
[ 496041986, 19249760]]) #random
Type: builtin_function_or_method
dtypeはarrayに格納するデータの型を指定するようです。例えば、以下のようにするとstr型のデータを所持するarrayを作ることができます。
x = np.empty([2, 2], dtype=str) x[0][0] = 'a' x[0][1] = 100 # 暗黙の型変換も行われます。 x
Output:
array([['a', '1'],
['', '']],
dtype='<U1')
dtype='<U1'は以下を参考に。ユニコードの1文字のデータ型のリトルエンディアンを表しているようです。リトル/ビッグエンディアンを何となくしか知らないので、これ以上の言及を避けておきます・・・
x = np.empty([2, 2])
デフォルトではdtype=floatなので、文字列を代入させることは出来ません。
x[0][0] = 'a'
Error:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-36-163567913498> in <module>
----> 1 x[0][0] = 'a'
ValueError: could not convert string to float: 'a'
dtype=objectの指定により、複数のオブジェクトを所持するarrayを作成することができます。
x = np.empty([2, 2], dtype=object) x[0][0] ='a' x[0][1] = 'b' x[1][0] = 3 x
Output:
array([['a', 'b'],
[3, None]], dtype=object)
例題:スペースシャトル「チャレンジャー号」の悲劇
makedirs("data", exist_ok=True) urlretrieve("https://git.io/vXknD", "data/challenger_data.csv")
Output:
('data/challenger_data.csv', <http.client.HTTPMessage at 0x1fadfa06198>)
figsize(25, 7) np.set_printoptions(precision=3, suppress=True) challenger_data = np.genfromtxt("data/challenger_data.csv", skip_header=1, usecols=[1, 2], missing_values="NA", delimiter=",") challenger_data
Output:
array([[ 66., 0.],
[ 70., 1.],
[ 69., 0.],
[ 80., nan],
[ 68., 0.],
[ 67., 0.],
[ 72., 0.],
[ 73., 0.],
[ 70., 0.],
[ 57., 1.],
[ 63., 1.],
[ 70., 1.],
[ 78., 0.],
[ 67., 0.],
[ 53., 1.],
[ 67., 0.],
[ 75., 0.],
[ 70., 0.],
[ 81., 0.],
[ 76., 0.],
[ 79., 0.],
[ 75., 1.],
[ 76., 0.],
[ 58., 1.],
[ 31., nan]])
challenger_data = challenger_data[~np.isnan(challenger_data[:, 1])]
challenger_data
Output:
array([[ 66., 0.],
[ 70., 1.],
[ 69., 0.],
[ 68., 0.],
[ 67., 0.],
[ 72., 0.],
[ 73., 0.],
[ 70., 0.],
[ 57., 1.],
[ 63., 1.],
[ 70., 1.],
[ 78., 0.],
[ 67., 0.],
[ 53., 1.],
[ 67., 0.],
[ 75., 0.],
[ 70., 0.],
[ 81., 0.],
[ 76., 0.],
[ 79., 0.],
[ 75., 1.],
[ 76., 0.],
[ 58., 1.]])
メモ
challenger_data[~np.isnan(challenger_data[:, 1])]の補足をしておきます。
challenger_data = np.genfromtxt("data/challenger_data.csv", skip_header=1, usecols=[1, 2], missing_values="NA", delimiter=",")
まず、2列目の成分を[:, 1]により取り出します。
challenger_data[:, 1]
Output
array([ 0., 1., 0., nan, 0., 0., 0., 0., 0., 1., 1.,
1., 0., 0., 1., 0., 0., 0., 0., 0., 0., 1.,
0., 1., nan])
np.isnanは入力されたarrayオブジェクトに対して、入力されたarrayオブジェクトの成分がnanならばTrueを、nan以外ならばFalseを所持するarrayオブジェクトを返します。
np.isnan??(クリックすると展開されます)
Call signature: np.isnan(*args, **kwargs)
Type: ufunc
String form: <ufunc 'isnan'>
File: c:\users\hoge\anaconda3\lib\site-packages\numpy\__init__.py
Docstring:
isnan(x[, out])
Test element-wise for NaN and return result as a boolean array.
Parameters
----------
x : array_like
Input array.
Returns
-------
y : ndarray or bool
For scalar input, the result is a new boolean with value True if
the input is NaN; otherwise the value is False.
For array input, the result is a boolean array of the same
dimensions as the input and the values are True if the
corresponding element of the input is NaN; otherwise the values are
False.
See Also
--------
isinf, isneginf, isposinf, isfinite
Notes
-----
Numpy uses the IEEE Standard for Binary Floating-Point for Arithmetic
(IEEE 754). This means that Not a Number is not equivalent to infinity.
Examples
--------
>>> np.isnan(np.nan)
True
>>> np.isnan(np.inf)
False
>>> np.isnan([np.log(-1.),1.,np.log(0)])
array([ True, False, False], dtype=bool)
Class docstring:
Functions that operate element by element on whole arrays.
To see the documentation for a specific ufunc, use np.info(). For
example, np.info(np.sin). Because ufuncs are written in C
(for speed) and linked into Python with NumPy's ufunc facility,
Python's help() function finds this page whenever help() is called
on a ufunc.
A detailed explanation of ufuncs can be found in the "ufuncs.rst"
file in the NumPy reference guide.
Unary ufuncs:
=============
op(X, out=None)
Apply op to X elementwise
Parameters
----------
X : array_like
Input array.
out : array_like
An array to store the output. Must be the same shape as `X`.
Returns
-------
r : array_like
`r` will have the same shape as `X`; if out is provided, `r`
will be equal to out.
Binary ufuncs:
==============
op(X, Y, out=None)
Apply `op` to `X` and `Y` elementwise. May "broadcast" to make
the shapes of `X` and `Y` congruent.
The broadcasting rules are:
* Dimensions of length 1 may be prepended to either array.
* Arrays may be repeated along dimensions of length 1.
Parameters
----------
X : array_like
First input array.
Y : array_like
Second input array.
out : array_like
An array to store the output. Must be the same shape as the
output would have.
Returns
-------
r : array_like
The return value; if out is provided, `r` will be equal to out.
例えば、先ほどの例では、以下のようなarrayオブジェクトが返されます。
np.isnan(challenger_data[:, 1])
Output:
array([False, False, False, True, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, True], dtype=bool)
そして、~によりブール値を反転させています。~は整数型のビットを反転させる演算子です。
参考:整数型におけるビット単位演算、符号付数値表現 の「2の補数」
~np.isnan(challenger_data[:, 1])
Output:
array([ True, True, True, False, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, False], dtype=bool)
最後に、~np.isnan(challenger_data[:, 1])の成分がTrueの行だけをスライスにより取り出します。
challenger_data = challenger_data[~np.isnan(challenger_data[:, 1])]
challenger_data
Output:
array([[ 66., 0.],
[ 70., 1.],
[ 69., 0.],
[ 68., 0.],
[ 67., 0.],
[ 72., 0.],
[ 73., 0.],
[ 70., 0.],
[ 57., 1.],
[ 63., 1.],
[ 70., 1.],
[ 78., 0.],
[ 67., 0.],
[ 53., 1.],
[ 67., 0.],
[ 75., 0.],
[ 70., 0.],
[ 81., 0.],
[ 76., 0.],
[ 79., 0.],
[ 75., 1.],
[ 76., 0.],
[ 58., 1.]])
補足は以上です。
plt.scatter(challenger_data[:, 0], challenger_data[:, 1], s=75, color="k", alpha=0.5) plt.yticks([0, 1]) plt.ylabel("Damage incident?") plt.xlabel("Outside temperature (Fahrenheit)") plt.title("Defects of the space shuttle O-rings versus temperature")
Output:
Text(0.5, 1.0, 'Defects of the space shuttle O-rings versus temperature')

figsize(24, 6) def logistic(x, beta, alpha=0): return 1.0 / (1.0 + np.exp(np.dot(beta, x) + alpha)) x = np.linspace(-4, 4, 100)
def plot_logistics(x=x, beta=[1, 3, 5], alpha=[1, -2, 7]): for b in beta: plt.plot(x, logistic(x, b), label=fr"$\beta = {b}$", ls="--", lw=1) for b, a in zip(beta, alpha): plt.plot(x, logistic(x, b, a), label=fr"$\beta = {b}, \alpha = {a}$") plt.title("Logistic functions for " r"different $\beta$ and $\alpha$ values") plt.xlabel("$x$") plt.ylabel("Logistic function at $x$") plt.legend(loc="lower left")
plot_logistics()

2.2.11 正規分布
temperature = challenger_data[:, 0] D = challenger_data[:, 1] beta = pm.Normal("beta", 0, 0.001, value=0) alpha = pm.Normal("alpha", 0, 0.001, value=0) @pm.deterministic def p(t=temperature, alpha=alpha, beta=beta): return 1.0 / (1. + np.exp(beta * t + alpha)) p.value # betaとalphaを定義したときにvalueを0に指定したのでpの各成分は0.5になっている。
Output:
array([ 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5,
0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5,
0.5])
observed = pm.Bernoulli("bernoulli_obs", p, value=D, observed=True) model = pm.Model([observed, beta, alpha]) map_ = pm.MAP(model) map_.fit() mcmc=pm.MCMC(model) mcmc.sample(120000, 100000, 2)
Output:
[-----------------100%-----------------] 120000 of 120000 complete in 11.2 sec
figsize(25, 12) alpha_samples = mcmc.trace('alpha')[:, None] beta_samples = mcmc.trace('beta')[:, None] plt.subplot(211) plt.title("Posterior distributions of " r"the model parameters $\alpha, \beta$") plt.hist(beta_samples, histtype='stepfilled', bins=35, alpha=0.85, color="#7A68A6", density=True, label=r"posterior of $\beta$") plt.legend() plt.subplot(212) plt.hist(alpha_samples, histtype='stepfilled', bins=35, alpha=0.85, color="#A60628", density=True, label=r"posterior of $\alpha$") plt.xlabel("Value of parameter") plt.ylabel("Density") plt.legend()
Output:
<matplotlib.legend.Legend at 0x1fae1ba67b8>

figsize(25, 8) t = np.linspace(temperature.min() - 5, temperature.max() + 5, 50)[:, None] p_t = logistic(t.T, beta_samples, alpha_samples) p_t
Output:
array([[ 0.998, 0.998, 0.997, ..., 0. , 0. , 0. ],
[ 0.998, 0.998, 0.997, ..., 0. , 0. , 0. ],
[ 1. , 0.999, 0.999, ..., 0.001, 0.001, 0.001],
...,
[ 0.989, 0.986, 0.983, ..., 0.003, 0.002, 0.002],
[ 0.989, 0.986, 0.983, ..., 0.003, 0.002, 0.002],
[ 0.987, 0.984, 0.98 , ..., 0.002, 0.002, 0.002]])
10000のサンプルが得られました。
p_t.shape
Output:
(10000, 50)
def plot_post_exp_val_of_prob_of_def(plot_real_list=[0, -2], fig_size_y=8, legend_flag=True, mean_lw=1): figsize(25, fig_size_y) mean_prob_t = p_t.mean(axis=0) for i in plot_real_list: plt.plot(t, p_t[i, :], ls="--", label=f"realization from posterior: {i}") plt.plot(t, mean_prob_t, lw=mean_lw, # plotするデータが多くなっても目立つようにlwを変更できるようにした。 color='r', label="average posterior \n" "probability of defect") plt.scatter(temperature, D, color="K", s=50, alpha=0.5) plt.title("Posterior expected value of the probability of defect, " f"including {len(plot_real_list)} realizations") if legend_flag: plt.legend(loc="lower left") plt.ylim(-0.1, 1.1) plt.xlim(t.min(), t.max()) plt.ylabel("Probability") plt.xlabel("Temperature")
plot_post_exp_val_of_prob_of_def()

10000のデータから100個のデータを100間隔でプロットしてみました。いろんな値がサンプリングされているということがわかりますね。
plot_post_exp_val_of_prob_of_def(plot_real_list=[i for i in range(0, 10000, 100)], fig_size_y=8, legend_flag=False)

qs = mquantiles(p_t, [0.025, 0.975], axis=0) figsize(25, 8) mean_prob_t = p_t.mean(axis=0) plt.fill_between(t[:, 0], *qs, alpha=0.7, color="#7A68A6") plt.plot(t[:, 0], qs[0], label="95% CI", color="#7A68A6", alpha=0.7) plt.plot(t, mean_prob_t, lw=1, ls="--", color="k", label="average posterior \nprobability of defect") plt.xlim(t.min(), t.max()) plt.ylim(-0.02, 1.02) plt.legend(loc="lower left") plt.scatter(temperature, D, color="k", s=50, alpha=0.5) plt.xlabel("Temprature, $t$") plt.ylabel("Probability estimate") plt.title("Posterior probability of estimates, given temperature $t$")
Output:
Text(0.5, 1.0, 'Posterior probability of estimates, given temperature $t$')

2.2.12 チャレンジャー号の悲劇の日に何が起こった?
figsize(25, 5) prob_31 = logistic(31, beta_samples, alpha_samples) plt.hist(prob_31, bins=1000, density=True, histtype='stepfilled') plt.xlim(0.995, 1) plt.ylabel("Density") plt.xlabel("Probability of defect occurring in O-ring") plt.title("Posterior distribution of probability of defect, " "given $t = 31")
Output:
Text(0.5, 1.0, 'Posterior distribution of probability of defect, given $t = 31')

2.3 このモデルは適切か?
simulated = pm.Bernoulli("bernoulli_sim", p) N = 10000 mcmc = pm.MCMC([simulated, alpha, beta, observed]) mcmc.sample(N)
Output:
[-----------------100%-----------------] 10000 of 10000 complete in 1.9 sec
simulations = mcmc.trace("bernoulli_sim")[:].astype(int)
メモ
.astype(int)の補足。mcmc.trace("bernoulli_sim")[:]のdtypeはbool値となっています。
mcmc.trace("bernoulli_sim")[:]
Output:
array([[False, True, False, ..., False, True, True],
[False, False, False, ..., False, False, True],
[False, False, False, ..., False, False, True],
...,
[ True, False, False, ..., False, False, True],
[ True, False, False, ..., False, False, True],
[False, False, False, ..., False, False, True]], dtype=bool)
そこで、astypeメソッドを使って、整数型に変換しています。
mcmc.trace("bernoulli_sim")[:].astype(int)
Output:
array([[0, 1, 0, ..., 0, 1, 1],
[0, 0, 0, ..., 0, 0, 1],
[0, 0, 0, ..., 0, 0, 1],
...,
[1, 0, 0, ..., 0, 0, 1],
[1, 0, 0, ..., 0, 0, 1],
[0, 0, 0, ..., 0, 0, 1]])
以上、メモでした。
simulations.shape
Output:
(10000, 23)
plt.suptitle("Simulated datasets using posterior parameters") figsize(25, 20) for i in range(4): ax = plt.subplot(4, 1, i + 1) plt.scatter(temperature, simulations[1000 * i, :], color="k", s=50, alpha=0.5)
Output:

2.3.1 セパレーションプロット
posterior_probability = simulations.mean(axis=0) print("Obs. | Arraya of Simulated Defects | posteriror | Realized") print(" Probability Defect ") print(" of Defect ") for i in range(len(D)): print(f'{str(i).zfill(2)} | {str(simulations[:10, i])[:-1] + "...]".ljust(6)} | {round(posterior_probability[i], 2)} | {int(D[i])}')
Output:
Obs. | Arraya of Simulated Defects | posteriror | Realized
Probability Defect
of Defect
00 | [0 0 0 0 0 1 1 0 0 0...] | 0.39 | 0
01 | [1 0 0 0 0 0 0 0 0 0...] | 0.15 | 1
02 | [0 0 0 0 0 0 0 0 0 0...] | 0.19 | 0
03 | [1 0 1 0 0 1 0 0 0 1...] | 0.26 | 0
04 | [0 0 0 0 1 0 0 0 1 0...] | 0.31 | 0
05 | [0 0 0 0 0 0 0 0 1 0...] | 0.08 | 0
06 | [0 0 0 0 1 0 0 0 0 0...] | 0.06 | 0
07 | [0 0 0 0 1 0 0 0 0 0...] | 0.14 | 0
08 | [1 1 1 1 0 1 1 1 1 1...] | 0.92 | 1
09 | [0 0 0 1 0 1 1 1 1 1...] | 0.63 | 1
10 | [0 0 0 0 0 0 0 0 0 0...] | 0.14 | 1
11 | [0 0 0 0 1 0 0 0 0 0...] | 0.01 | 0
12 | [1 0 0 0 0 0 0 0 0 0...] | 0.32 | 0
13 | [0 1 1 0 1 1 1 1 1 1...] | 0.97 | 1
14 | [0 1 1 0 0 0 1 1 0 0...] | 0.31 | 0
15 | [0 0 0 0 0 0 0 0 0 0...] | 0.03 | 0
16 | [0 0 0 0 0 0 0 0 0 0...] | 0.14 | 0
17 | [0 0 0 0 0 0 0 0 0 0...] | 0.01 | 0
18 | [0 0 0 0 0 0 0 0 0 0...] | 0.03 | 0
19 | [0 0 0 0 0 0 0 0 0 0...] | 0.01 | 0
20 | [0 0 0 0 0 0 0 0 0 0...] | 0.03 | 1
21 | [1 0 0 0 0 0 0 0 0 0...] | 0.03 | 0
22 | [1 1 1 0 1 1 1 1 1 1...] | 0.89 | 1
ix = np.argsort(posterior_probability) print("Posterior Probability of Defect | Realized Defect") for i in range(len(D)): print(f"{round(posterior_probability[ix[i]], 2)} | {int(D[ix[i]])}")
Output:
Posterior Probability of Defect | Realized Defect
0.01 | 0
0.01 | 0
0.01 | 0
0.03 | 0
0.03 | 0
0.03 | 1
0.03 | 0
0.06 | 0
0.08 | 0
0.14 | 0
0.14 | 0
0.14 | 1
0.15 | 1
0.19 | 0
0.26 | 0
0.31 | 0
0.31 | 0
0.32 | 0
0.39 | 0
0.63 | 1
0.89 | 1
0.92 | 1
0.97 | 1
urlretrieve("https://git.io/vXtye", 'separation_plot.py')
Output:
('separation_plot.py', <http.client.HTTPMessage at 0x1fae3e2ca58>)
from separation_plot import separation_plot figsize(22, 3) separation_plot(posterior_probability, D)

posterior_probability.shape[0]
Output:
23
figsize(22, 2.5) p = D separation_plot(p, D) plt.title("Perfect model") p = np.random.rand(23) separation_plot(p, D) plt.title("Random model") constant_prob = 7. / 23 * np.ones(23) separation_plot(constant_prob, D) plt.title("Constant-prediction model")
Output:
Text(0.5, 1.0, 'Constant-prediction model')



今回の写経は以上です。 ここまで読んでいただきありがとうございました。
Pythonで体験するベイズ推論 PyMCによるMCMC入門 第2章 (2.2.4まで) 写経
始めに
この記事では、Pythonで体験するベイズ推論 PyMCによるMCMC入門
の第2章の2.2.4までを写経した記録をまとめます。前回はこちらPythonで体験するベイズ推論 PyMCによるMCMC入門 第1章 写経。 実行はJupyter Labにて実行しています。
写経は以下のようにまとめました。
写経ではありますが、コードを実行しやすいように関数を作成し、コードの再実行がやりやすいようにした部分があります。
また、より良いと思われるコードを考えた場合は書き換えてコメントを添えるようにし、変更点をなるべく明示するようにしてあります。
個人的に気になった点のメモを残すようにしてあります。同じような疑問を持った方の助けになれば幸いです。
以前書いたコードと同じようなコード(例えばグラフの描写等)は効率化のために飛ばしているところもあります。
Pythonで体験するベイズ推論 PyMCによるMCMC入門を読んでいている際のちょっとした正誤表代わりになればと思います。
2 PyMCについてもう少し
2.1 はじめに
2.1.1 親子関係
import pymc as pm
lambda_ = pm.Exponential("poisson_param", 1) data_generator = pm.Poisson("data_generator", lambda_) data_plus_one = data_generator + 1
print("Childrem of lambda_: ") print(lambda_.children, "\n") print("Parents of data_generator: ") print(data_generator.parents, "\n") print("Children of data_generator: ") print(data_generator.children)
Output:
Childrem of lambda_:
{<pymc.distributions.new_dist_class.<locals>.new_class 'data_generator' at 0x000002720195BB38>}
Parents of data_generator:
{'mu': <pymc.distributions.new_dist_class.<locals>.new_class 'poisson_param' at 0x000002720195BB00>}
Children of data_generator:
{<pymc.PyMCObjects.Deterministic '(data_generator_add_1)' at 0x000002720195BBA8>}
2.1.2 PyMC変数
print(f"lambda_.value = {lambda_.value}") print(f"data_generator.value = {data_generator.value}") print(f"data_plus_one.value = {data_plus_one.value}")
Output:
lambda_.value = 0.10977163451249013
data_generator.value = 0
data_plus_one.value = 1
discrete_uni_var = pm.DiscreteUniform("discrete_uni_var", 0, 4)
discrete_uni_varの説明を確認しておきます。
discrete_uni_var??
Output:
Signature: discrete_uni_var(*args, **kwargs)
Type: DiscreteUniform
String form: discrete_uni_var
File: c:\users\hoge\anaconda3\lib\site-packages\pymc\distributions.py
Docstring: <no docstring>
Class docstring:
D = DiscreteUniform(name, lower, upper, value=None, observed=False, size=1, trace=True, rseed=True, doc=None, verbose=-1, debug=False)
Stochastic variable with DiscreteUniform distribution.
Parents are: lower, upper.
Docstring of log-probability function:
Discrete uniform log-likelihood.
.. math::
f(x \mid lower, upper) = \frac{1}{upper-lower}
:Parameters:
- `x` : [int] :math:`lower \leq x \leq upper`
- `lower` : Lower limit.
- `upper` : Upper limit (upper > lower).
betas = pm.Uniform("betas", 0, 1, size=10) betas.value
Output:
array([ 0.34388063, 0.71044819, 0.8913627 , 0.19832807, 0.57029206,
0.5054775 , 0.55222677, 0.12595211, 0.55920651, 0.34046255])
lambda_1 = pm.Exponential("lambda_1", 1) lambda_2 = pm.Exponential("lambda_2", 1) tau = pm.DiscreteUniform("tau", lower=0, upper=10) print("Initialized values...") print(f"lambda_1.value = {lambda_1.value}") print(f"lambda_2.value = {lambda_2.value}") print(f"tau.value = {tau.value}\n") lambda_1.random() lambda_2.random() tau.random() print("After calling random() on the variables...") print(f"lambda_1.value = {lambda_1.value}") print(f"lambda_2.value = {lambda_2.value}") print(f"tau.value = {tau.value}")
Output:
Initialized values...
lambda_1.value = 0.08344212711504026
lambda_2.value = 0.009108273929809445
tau.value = 10
After calling random() on the variables...
lambda_1.value = 0.33157006248423776
lambda_2.value = 0.46469007162531084
tau.value = 6
type(lambda_1 + lambda_2)
pymc.PyMCObjects.Deterministic
@pm.deterministic def some_deterministic(stoch=discrete_uni_var): return stoch.value**2 # valueを付けるとエラー
Error:
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-10-a9bb7333fdfc> in <module>
1 @pm.deterministic
----> 2 def some_deterministic(stoch=discrete_uni_var):
3 return stoch.value**2 # valueを付けるとエラー
~\Anaconda3\lib\site-packages\pymc\InstantiationDecorators.py in deterministic(__func__, **kwds)
248
249 if __func__:
--> 250 return instantiate_n(__func__)
251
252 return instantiate_n
~\Anaconda3\lib\site-packages\pymc\InstantiationDecorators.py in instantiate_n(__func__)
241 junk, parents = _extract(
242 __func__, kwds, keys, 'Deterministic', probe=False)
--> 243 return Deterministic(parents=parents, **kwds)
244
245 keys = ['eval']
~\Anaconda3\lib\site-packages\pymc\PyMCObjects.py in __init__(self, eval, doc, name, parents, dtype, trace, cache_depth, plot, verbose, jacobians, jacobian_formats)
441 trace=trace,
442 plot=plot,
--> 443 verbose=verbose)
444
445 # self._value.force_compute()
~\Anaconda3\lib\site-packages\pymc\Node.py in __init__(self, doc, name, parents, cache_depth, trace, dtype, plot, verbose)
212 self.extended_children = set()
213
--> 214 Node.__init__(self, doc, name, parents, cache_depth, verbose=verbose)
215
216 if self.dtype is None:
~\Anaconda3\lib\site-packages\pymc\Node.py in __init__(self, doc, name, parents, cache_depth, verbose)
127
128 # Initialize
--> 129 self.parents = parents
130
131 def _get_parents(self):
~\Anaconda3\lib\site-packages\pymc\Node.py in _set_parents(self, new_parents)
145
146 # Get new lazy function
--> 147 self.gen_lazy_function()
148
149 parents = property(
~\Anaconda3\lib\site-packages\pymc\PyMCObjects.py in gen_lazy_function(self)
452 cache_depth=self._cache_depth)
453
--> 454 self._value.force_compute()
455
456 self._jacobians = {}
LazyFunction.pyx in pymc.LazyFunction.LazyFunction.force_compute (pymc\LazyFunction.c:2409)()
<ipython-input-10-a9bb7333fdfc> in some_deterministic(stoch)
1 @pm.deterministic
2 def some_deterministic(stoch=discrete_uni_var):
----> 3 return stoch.value**2 # valueを付けるとエラー
AttributeError: 'numpy.ndarray' object has no attribute 'value'
@pm.deterministic def some_deterministic(stoch=discrete_uni_var): return stoch**2
@pm.deterministic def some_deterministic(stoch): # deterministic関数の引数がキーワードとして渡されていないのでエラー return stoch**2
Error:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-12-e91cec33d01b> in <module>
----> 1 @pm.deterministic
2 def some_deterministic(stoch): # deterministic関数の引数がキーワードとして渡されていないのでエラー
3 return stoch**2
~\Anaconda3\lib\site-packages\pymc\InstantiationDecorators.py in deterministic(__func__, **kwds)
248
249 if __func__:
--> 250 return instantiate_n(__func__)
251
252 return instantiate_n
~\Anaconda3\lib\site-packages\pymc\InstantiationDecorators.py in instantiate_n(__func__)
240 def instantiate_n(__func__):
241 junk, parents = _extract(
--> 242 __func__, kwds, keys, 'Deterministic', probe=False)
243 return Deterministic(parents=parents, **kwds)
244
~\Anaconda3\lib\site-packages\pymc\InstantiationDecorators.py in _extract(__func__, kwds, keys, classname, probe)
113 if i < arg_deficit - 1:
114 err_str += ','
--> 115 raise ValueError(err_str)
116
117 # Fill in parent dictionary
ValueError: Deterministic some_deterministic: no parent provided for the following labels: stoch
2.1.3 モデルに観測を組み込む
from IPython.core.pylabtools import figsize from matplotlib import pyplot as plt %matplotlib inline import numpy as np def plot_prior_dist_for_lambda_1(sample_size=20000): figsize(25, 8) plt.rcParams['font.size'] = 17 # フォントのサイズ指定する。 samples = [lambda_1.random() for i in range(sample_size)] plt.hist(samples, bins=70, density=True, histtype="stepfilled") plt.title("Prior distribution for $\lambda_1$") plt.xlabel("Value") plt.ylabel("Density") plt.xlim(0, 8)
plot_prior_dist_for_lambda_1()

メモ
初めて読んだときはは何言ってるかわかりませんでしたがvalue=dataと共にobserved=Trueを指定して、pm.Poissonを呼び出したら、random()の呼び出しに関わらずポアソン分布ではなくてvalueに渡した観測値が毎回返ってくるということを示したかったかな?そういう意味ではポアソン分布である必要はない気がする。
data = np.array([10, 5]) fixed_variable = pm.Poisson("fxd", 1, value=data, observed=True) print(f"value: {fixed_variable.value}") # valueの表示 print("calling .random()") # .random()の呼び出し fixed_variable.random() print(f"value: {fixed_variable.value}")
Output:
value: [10 5]
calling .random()
value: [10 5]
指数分布を使って同様のコードを実行してみました。この場合はvalueで返される値がfloat型になる以外はポアソン分布のときと同じ結果のよう。
data = np.array([10, 5]) fixed_variable = pm.Exponential("fxd", 1, value=data, observed=True) print(f"value: {fixed_variable.value}") # valueの表示 print("calling .random()") # .random()の呼び出し fixed_variable.random() print(f"value: {fixed_variable.value}")
Output:
value: [ 10. 5.]
calling .random()
value: [ 10. 5.]
これだったら、初めから観測値の分布を表すためのクラスが作ってあっても良いんじゃないかな?と思う.ただ、ある分布から得られた観測値ということを表現することが大事なのかもしれない。例えば、この場合は平均1の指数分布からの観測値が10と5だったというように。こればかりは今後の説明を待つしかないかもしれませんね。
data = np.array([10, 25, 15, 20, 35]) obs = pm.Poisson("obs", lambda_, value=data, observed=True) print(obs.value)
Output:
[10 25 15 20 35]
2.1.4 最後に
model = pm.Model([obs, lambda_, lambda_1, lambda_2, tau])
2.2 モデリングのアプローチ
2.2.1 同じ物語、異なる結末
tau = pm.rdiscrete_uniform(0, 80) print(tau)
Output:
50
alpha = 1 / 20 lambda_1, lambda_2 = pm.rexponential(alpha, 2) #引数の2はサイズ。 print(lambda_1, lambda_2)
Output:
3.31182568691 11.2489589986
lambda_ = np.r_[lambda_1 * np.ones(tau),
lambda_2 * np.ones(80 - tau)]
print(lambda_)
Output:
[ 3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
3.31182569 3.31182569 3.31182569 3.31182569 3.31182569
11.248959 11.248959 11.248959 11.248959 11.248959 11.248959
11.248959 11.248959 11.248959 11.248959 11.248959 11.248959
11.248959 11.248959 11.248959 11.248959 11.248959 11.248959
11.248959 11.248959 11.248959 11.248959 11.248959 11.248959
11.248959 11.248959 11.248959 11.248959 11.248959 11.248959 ]
メモ
np.r_はここでは二つのarrayオブジェクトを結合させるために使っているようです。
ただ、それ以上の使い方を読もうとしてもnumpy自体に疎いので説明が全く理解できませんでした…
help(np.lib.index_tricks.r_)
Output:
Help on RClass in module numpy.lib.index_tricks object:
class RClass(AxisConcatenator)
| Translates slice objects to concatenation along the first axis.
|
| This is a simple way to build up arrays quickly. There are two use cases.
|
| 1. If the index expression contains comma separated arrays, then stack
| them along their first axis.
| 2. If the index expression contains slice notation or scalars then create
| a 1-D array with a range indicated by the slice notation.
|
| If slice notation is used, the syntax ``start:stop:step`` is equivalent
| to ``np.arange(start, stop, step)`` inside of the brackets. However, if
| ``step`` is an imaginary number (i.e. 100j) then its integer portion is
| interpreted as a number-of-points desired and the start and stop are
| inclusive. In other words ``start:stop:stepj`` is interpreted as
| ``np.linspace(start, stop, step, endpoint=1)`` inside of the brackets.
| After expansion of slice notation, all comma separated sequences are
| concatenated together.
|
| Optional character strings placed as the first element of the index
| expression can be used to change the output. The strings 'r' or 'c' result
| in matrix output. If the result is 1-D and 'r' is specified a 1 x N (row)
| matrix is produced. If the result is 1-D and 'c' is specified, then a N x 1
| (column) matrix is produced. If the result is 2-D then both provide the
| same matrix result.
|
| A string integer specifies which axis to stack multiple comma separated
| arrays along. A string of two comma-separated integers allows indication
| of the minimum number of dimensions to force each entry into as the
| second integer (the axis to concatenate along is still the first integer).
|
| A string with three comma-separated integers allows specification of the
| axis to concatenate along, the minimum number of dimensions to force the
| entries to, and which axis should contain the start of the arrays which
| are less than the specified number of dimensions. In other words the third
| integer allows you to specify where the 1's should be placed in the shape
| of the arrays that have their shapes upgraded. By default, they are placed
| in the front of the shape tuple. The third argument allows you to specify
| where the start of the array should be instead. Thus, a third argument of
| '0' would place the 1's at the end of the array shape. Negative integers
| specify where in the new shape tuple the last dimension of upgraded arrays
| should be placed, so the default is '-1'.
|
| Parameters
| ----------
| Not a function, so takes no parameters
|
|
| Returns
| -------
| A concatenated ndarray or matrix.
|
| See Also
| --------
| concatenate : Join a sequence of arrays along an existing axis.
| c_ : Translates slice objects to concatenation along the second axis.
|
| Examples
| --------
| >>> np.r_[np.array([1,2,3]), 0, 0, np.array([4,5,6])]
| array([1, 2, 3, 0, 0, 4, 5, 6])
| >>> np.r_[-1:1:6j, [0]*3, 5, 6]
| array([-1. , -0.6, -0.2, 0.2, 0.6, 1. , 0. , 0. , 0. , 5. , 6. ])
|
| String integers specify the axis to concatenate along or the minimum
| number of dimensions to force entries into.
|
| >>> a = np.array([[0, 1, 2], [3, 4, 5]])
| >>> np.r_['-1', a, a] # concatenate along last axis
| array([[0, 1, 2, 0, 1, 2],
| [3, 4, 5, 3, 4, 5]])
| >>> np.r_['0,2', [1,2,3], [4,5,6]] # concatenate along first axis, dim>=2
| array([[1, 2, 3],
| [4, 5, 6]])
|
| >>> np.r_['0,2,0', [1,2,3], [4,5,6]]
| array([[1],
| [2],
| [3],
| [4],
| [5],
| [6]])
| >>> np.r_['1,2,0', [1,2,3], [4,5,6]]
| array([[1, 4],
| [2, 5],
| [3, 6]])
|
| Using 'r' or 'c' as a first string argument creates a matrix.
|
| >>> np.r_['r',[1,2,3], [4,5,6]]
| matrix([[1, 2, 3, 4, 5, 6]])
|
| Method resolution order:
| RClass
| AxisConcatenator
| builtins.object
|
| Methods defined here:
|
| __init__(self)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Methods inherited from AxisConcatenator:
|
| __getitem__(self, key)
|
| __getslice__(self, i, j)
|
| __len__(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from AxisConcatenator:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
data = pm.rpoisson(lambda_)
print(data)
Output:
[ 3 6 5 1 3 3 2 2 0 4 3 4 2 2 4 1 1 3 2 1 4 4 6 3 9
4 4 1 5 2 1 6 2 7 5 2 1 2 3 3 3 7 3 5 2 2 8 3 4 7
10 6 12 13 8 13 9 19 10 16 9 11 11 12 9 9 8 18 11 9 7 15 5 11 12
8 16 12 9 10]
plt.bar(np.arange(80), data, color="#348ABD") plt.bar(tau - 1, data[tau - 1], color="r", label="user behavior changed") plt.xlabel("Time (days)") plt.ylabel("Text messages received") plt.title("Artificial dataset from simulating the model") plt.xlim(0, 80) plt.legend()
<matplotlib.legend.Legend at 0x27207f35908>

def plot_artificial_sms_dataset(): tau = pm.rdiscrete_uniform(0, 80) alpha = 1. / 20. lambda_1, lambda_2 = pm.rexponential(alpha, 2) data = np.r_[pm.rpoisson(lambda_1, tau), pm.rpoisson(lambda_2, 80 - tau)] plt.bar(np.arange(80), data, color="#348ABD") plt.bar(tau - 1, data[tau - 1], color="r", label="user behavior changed") plt.xlim(0, 80) figsize(25, 10) plt.suptitle("More examples of artificial datasets " "from simulating our model") for i in range(4): plt.subplot(4, 1, 1 + i) plt.xlabel("Time (days)") plot_artificial_sms_dataset()

2.2.3 単純な場合
p = pm.Uniform('p', lower=0, upper=1)
p_true = 0.05 N = 1500 occurrences = pm.rbernoulli(p_true, N) print(occurrences) print(occurrences.sum())
Output:
[False False False ..., False False False]
72
print(f"What is the observed frequency in Group A? {occurrences.mean()}") print(f"Does the observed frequency equal the true frequency? {occurrences.mean() == p_true}")
Output:
What is the observed frequency in Group A? 0.048
Does the observed frequency equal the true frequency? False
obs = pm.Bernoulli("obs", p, value=occurrences, observed=True) mcmc = pm.MCMC([p, obs]) mcmc.sample(20000, 1000) figsize(25, 8) plt.vlines(p_true, 0, 200, linestyles="--", # テキストでは90だったが、200に修正しておいた。どうも、テキストの図よりもヒストグラムの最大値が大きくなってしまう。 label="true $p_A$ (unknown)") plt.hist(mcmc.trace("p")[:], bins=35, histtype="stepfilled", density=True) plt.title("Posterior distribution of $p_A$, " "the true effectiveness of site A") plt.xlabel("Value of $p_A$") plt.ylabel("Density") plt.legend()
[-----------------100%-----------------] 20000 of 20000 complete in 9.3 sec
<matplotlib.legend.Legend at 0x27209ac7b70>

2.2.4 AとBを一緒に
true_p_A = 0.05 true_p_B = 0.04 N_A = 1500 N_B = 750 observations_A = pm.rbernoulli(true_p_A, N_A) observations_B = pm.rbernoulli(true_p_B, N_B) print(f"Obs from Site A: {observations_A[:30].astype(int)}...") print(f"Obs from Site B: {observations_B[:30].astype(int)}...")
Output:
Obs from Site A: [0 0 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]...
Obs from Site B: [0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0]...
print(observations_A.mean()) print(observations_B.mean())
Output:
0.0526666666667
0.0386666666667
p_A = pm.Uniform("p_A", 0, 1) p_B = pm.Uniform("p_B", 0, 1) @pm.deterministic def delta(p_A=p_A, p_B=p_B): return p_A - p_B obs_A = pm.Bernoulli("obs_A", p_A, value=observations_A, observed=True) obs_B = pm.Bernoulli("obs_B", p_B, value=observations_B, observed=True) mcmc = pm.MCMC([p_A, p_B, delta, obs_A, obs_B]) mcmc.sample(25000, 5000)
Ooutput:
[-----------------100%-----------------] 25000 of 25000 complete in 4.3 sec
p_A_samples = mcmc.trace("p_A")[:] p_B_samples = mcmc.trace("p_B")[:] delta_samples = mcmc.trace("delta")[:] figsize(25, 20) ax = plt.subplot(311) plt.hist(p_A_samples, histtype='stepfilled', bins=30, alpha=0.85, label="posterior of $p_A$", color="#A60628", density=True) plt.vlines(true_p_A, 0, 80, linestyles="--", label="true $p_A$ (unknown)") plt.legend(loc="upper right") plt.xlim(0, .1) plt.ylim(0, 80) plt.suptitle("Posterior distributions of " "$p_A$, $p_B$, and delta unknown") ax = plt.subplot(312) plt.hist(p_B_samples, histtype='stepfilled', bins=30, alpha=0.85, label="posterior of $p_B$", color="#467821", density=True) plt.vlines(true_p_B, 0, 80, linestyles="--", label="true $p_B$ (unknown)") plt.legend(loc="upper right") plt.xlim(0, .1) plt.ylim(0, 80) plt.ylabel("Density") ax = plt.subplot(313) plt.hist(delta_samples, histtype='stepfilled', bins=30, alpha=0.85, label="posterior of delta", color="#7A68A5", density=True) plt.vlines(true_p_A - true_p_B, 0, 60, linestyles="--", label="true delta (unknown)") plt.vlines(0, 0, 60, color="black", alpha=0.2) plt.xlabel("Value") plt.legend(loc="upper right")
<matplotlib.legend.Legend at 0x2720a2c90f0>

figsize(25, 6) plt.hist(p_A_samples, histtype='stepfilled', bins=30, alpha=0.80, label="posterior of $p_A$", color="#A60628", density=True) plt.hist(p_B_samples, histtype='stepfilled', bins=30, alpha=0.80, label="posterior of $p_B$", color="#467821", density=True) plt.legend(loc="upper right") plt.xlabel("Value") plt.ylabel("Density") plt.title("Posterior distributions of $p_A$ and $p_B$") plt.ylim(0, 80) plt.xlim(0, .1)
(0, 0.1)

print(f"Probability site A is WORSE tahn B: {(delta_samples < 0).mean()}") print(f"Probability site A is BETTER tahn B: {(delta_samples > 0).mean()}")
Output:
Probability site A is WORSE tahn B: 0.07845
Probability site A is BETTER tahn B: 0.92155
メモ
true_p_A、 true_p_B、 N_A、 N_Bを色々変えられるような関数を作ってみました。おもしろい。ただ、plt.vlinesのymax引数を動的に調整するにはどうすればいいのかな?と思った。
def plot_post_dist_p_A_p_B_delta(true_p_A=0.05, true_p_B=0.04, N_A=1500, N_B=750): true_p_A = true_p_A true_p_B = true_p_B N_A = N_A N_B = N_B observations_A = pm.rbernoulli(true_p_A, N_A) observations_B = pm.rbernoulli(true_p_B, N_B) p_A = pm.Uniform("p_A", 0, 1) p_B = pm.Uniform("p_B", 0, 1) @pm.deterministic def delta(p_A=p_A, p_B=p_B): return p_A - p_B obs_A = pm.Bernoulli("obs_A", p_A, value=observations_A, observed=True) obs_B = pm.Bernoulli("obs_B", p_B, value=observations_B, observed=True) mcmc = pm.MCMC([p_A, p_B, delta, obs_A, obs_B]) mcmc.sample(25000, 5000) p_A_samples = mcmc.trace("p_A")[:] p_B_samples = mcmc.trace("p_B")[:] delta_samples = mcmc.trace("delta")[:] figsize(25, 20) ax = plt.subplot(311) plt.hist(p_A_samples, histtype='stepfilled', bins=30, alpha=0.85, label="posterior of $p_A$", color="#A60628", density=True) plt.vlines(true_p_A, ymin=0, ymax=80, linestyles="--", label="true $p_A$ (unknown)") plt.legend(loc="upper right") plt.xlim(min(p_A_samples), max(p_A_samples)) # plt.ylim(0, 80) plt.suptitle("Posterior distributions of " "$p_A$, $p_B$, and delta unknown") ax = plt.subplot(312) plt.hist(p_B_samples, histtype='stepfilled', bins=30, alpha=0.85, label="posterior of $p_B$", color="#467821", density=True) plt.vlines(true_p_B, ymin=0, ymax=80, linestyles="--", label="true $p_B$ (unknown)") plt.legend(loc="upper right") plt.xlim(0, .1) # plt.ylim(0, 80) plt.ylabel("Density") ax = plt.subplot(313) plt.hist(delta_samples, histtype='stepfilled', bins=30, alpha=0.85, label="posterior of delta", color="#7A68A5", density=True) plt.vlines(true_p_A - true_p_B, 0, 60, linestyles="--", label="true delta (unknown)") plt.vlines(0, 0, 60, color="black", alpha=0.2) plt.xlabel("Value") plt.legend(loc="upper right")
plot_post_dist_p_A_p_B_delta(true_p_A=0.1, N_A=10000, N_B=10000)
[-----------------100%-----------------] 25000 of 25000 complete in 12.5 sec

今回の写経は以上です。 ここまで読んでいただきありがとうございました。
Numpyのgenfromtxtについて少しまとめておいた
概要
Numpyのgenfromtxt(ドキュメント:Importing data with genfromtxt)の簡単な使い方をまとめておきます。内容としてはPythonで体験するベイズ推論 PyMCによるMCMC入門の2章 2.2.10 例題:スペースシャトル「チャレンジャー号」の悲劇で使用されたgenfromtxtを例にしてまとめておきます。
解説
まず、使用されていたコードは以下のようなものでした。
import numpy as np challenger_data = np.genfromtxt("data/challenger_data.csv", skip_header=1, usecols=[1, 2], missing_values="NA", delimiter=",")
data/challenger_data.csvファイルは以下のような内容でした。
Date,Temperature,Damage Incident 04/12/1981,66,0 11/12/1981,70,1 3/22/82,69,0 6/27/82,80,NA 01/11/1982,68,0 04/04/1983,67,0 6/18/83,72,0 8/30/83,73,0 11/28/83,70,0 02/03/1984,57,1 04/06/1984,63,1 8/30/84,70,1 10/05/1984,78,0 11/08/1984,67,0 1/24/85,53,1 04/12/1985,67,0 4/29/85,75,0 6/17/85,70,0 7/29/85,81,0 8/27/85,76,0 10/03/1985,79,0 10/30/85,75,1 11/26/85,76,0 01/12/1986,58,1 1/28/86,31,Challenger Accident
このコードによって生成されるchallenger_dataは以下のようなarrayオブジェクトです。
challenger_data
Output:
array([[ 66., 0.],
[ 70., 1.],
[ 69., 0.],
[ 80., nan],
[ 68., 0.],
[ 67., 0.],
[ 72., 0.],
[ 73., 0.],
[ 70., 0.],
[ 57., 1.],
[ 63., 1.],
[ 70., 1.],
[ 78., 0.],
[ 67., 0.],
[ 53., 1.],
[ 67., 0.],
[ 75., 0.],
[ 70., 0.],
[ 81., 0.],
[ 76., 0.],
[ 79., 0.],
[ 75., 1.],
[ 76., 0.],
[ 58., 1.],
[ 31., nan]])
np.genfromtxtのヘルプ
まず、np.genfromtxtのヘルプを見てみます。
np.genfromtxt?
Output:
Signature:
np.genfromtxt(
fname,
dtype=<class 'float'>,
comments='#',
delimiter=None,
skip_header=0,
skip_footer=0,
converters=None,
missing_values=None,
filling_values=None,
usecols=None,
names=None,
excludelist=None,
deletechars=None,
replace_space='_',
autostrip=False,
case_sensitive=True,
defaultfmt='f%i',
unpack=None,
usemask=False,
loose=True,
invalid_raise=True,
max_rows=None,
)
Docstring:
Load data from a text file, with missing values handled as specified.
Each line past the first `skip_header` lines is split at the `delimiter`
character, and characters following the `comments` character are discarded.
Parameters
----------
fname : file, str, list of str, generator
File, filename, list, or generator to read. If the filename
extension is `.gz` or `.bz2`, the file is first decompressed. Mote
that generators must return byte strings in Python 3k. The strings
in a list or produced by a generator are treated as lines.
dtype : dtype, optional
Data type of the resulting array.
If None, the dtypes will be determined by the contents of each
column, individually.
comments : str, optional
The character used to indicate the start of a comment.
All the characters occurring on a line after a comment are discarded
delimiter : str, int, or sequence, optional
The string used to separate values. By default, any consecutive
whitespaces act as delimiter. An integer or sequence of integers
can also be provided as width(s) of each field.
skiprows : int, optional
`skiprows` was removed in numpy 1.10. Please use `skip_header` instead.
skip_header : int, optional
The number of lines to skip at the beginning of the file.
skip_footer : int, optional
The number of lines to skip at the end of the file.
converters : variable, optional
The set of functions that convert the data of a column to a value.
The converters can also be used to provide a default value
for missing data: ``converters = {3: lambda s: float(s or 0)}``.
missing : variable, optional
`missing` was removed in numpy 1.10. Please use `missing_values`
instead.
missing_values : variable, optional
The set of strings corresponding to missing data.
filling_values : variable, optional
The set of values to be used as default when the data are missing.
usecols : sequence, optional
Which columns to read, with 0 being the first. For example,
``usecols = (1, 4, 5)`` will extract the 2nd, 5th and 6th columns.
names : {None, True, str, sequence}, optional
If `names` is True, the field names are read from the first valid line
after the first `skip_header` lines.
If `names` is a sequence or a single-string of comma-separated names,
the names will be used to define the field names in a structured dtype.
If `names` is None, the names of the dtype fields will be used, if any.
excludelist : sequence, optional
A list of names to exclude. This list is appended to the default list
['return','file','print']. Excluded names are appended an underscore:
for example, `file` would become `file_`.
deletechars : str, optional
A string combining invalid characters that must be deleted from the
names.
defaultfmt : str, optional
A format used to define default field names, such as "f%i" or "f_%02i".
autostrip : bool, optional
Whether to automatically strip white spaces from the variables.
replace_space : char, optional
Character(s) used in replacement of white spaces in the variables
names. By default, use a '_'.
case_sensitive : {True, False, 'upper', 'lower'}, optional
If True, field names are case sensitive.
If False or 'upper', field names are converted to upper case.
If 'lower', field names are converted to lower case.
unpack : bool, optional
If True, the returned array is transposed, so that arguments may be
unpacked using ``x, y, z = loadtxt(...)``
usemask : bool, optional
If True, return a masked array.
If False, return a regular array.
loose : bool, optional
If True, do not raise errors for invalid values.
invalid_raise : bool, optional
If True, an exception is raised if an inconsistency is detected in the
number of columns.
If False, a warning is emitted and the offending lines are skipped.
max_rows : int, optional
The maximum number of rows to read. Must not be used with skip_footer
at the same time. If given, the value must be at least 1. Default is
to read the entire file.
.. versionadded:: 1.10.0
Returns
-------
out : ndarray
Data read from the text file. If `usemask` is True, this is a
masked array.
See Also
--------
numpy.loadtxt : equivalent function when no data is missing.
Notes
-----
* When spaces are used as delimiters, or when no delimiter has been given
as input, there should not be any missing data between two fields.
* When the variables are named (either by a flexible dtype or with `names`,
there must not be any header in the file (else a ValueError
exception is raised).
* Individual values are not stripped of spaces by default.
When using a custom converter, make sure the function does remove spaces.
References
----------
.. [1] Numpy User Guide, section `I/O with Numpy
<http://docs.scipy.org/doc/numpy/user/basics.io.genfromtxt.html>`_.
Examples
---------
>>> from io import StringIO
>>> import numpy as np
Comma delimited file with mixed dtype
>>> s = StringIO("1,1.3,abcde")
>>> data = np.genfromtxt(s, dtype=[('myint','i8'),('myfloat','f8'),
... ('mystring','S5')], delimiter=",")
>>> data
array((1, 1.3, 'abcde'),
dtype=[('myint', '<i8'), ('myfloat', '<f8'), ('mystring', '|S5')])
Using dtype = None
>>> s.seek(0) # needed for StringIO example only
>>> data = np.genfromtxt(s, dtype=None,
... names = ['myint','myfloat','mystring'], delimiter=",")
>>> data
array((1, 1.3, 'abcde'),
dtype=[('myint', '<i8'), ('myfloat', '<f8'), ('mystring', '|S5')])
Specifying dtype and names
>>> s.seek(0)
>>> data = np.genfromtxt(s, dtype="i8,f8,S5",
... names=['myint','myfloat','mystring'], delimiter=",")
>>> data
array((1, 1.3, 'abcde'),
dtype=[('myint', '<i8'), ('myfloat', '<f8'), ('mystring', '|S5')])
An example with fixed-width columns
>>> s = StringIO("11.3abcde")
>>> data = np.genfromtxt(s, dtype=None, names=['intvar','fltvar','strvar'],
... delimiter=[1,3,5])
>>> data
array((1, 1.3, 'abcde'),
dtype=[('intvar', '<i8'), ('fltvar', '<f8'), ('strvar', '|S5')])
File: c:\users\hoge\anaconda3\lib\site-packages\numpy\lib\npyio.py
Type: function
全ての引数を説明することは難しいので、上のコードで使用されていた引数を以下ではまとめておきます。
fname引数
まず、fname引数についてみていきます。fnameはファイル名を指定するということは例のコードからもわかります。
また、fileオブジェクトのように、readメソッドで文字列を返すオブジェクトを渡すこともできます。
参考:Importing data with genfromtxt
例えば、以下のようにしても同じ結果が得られます。こんな面倒な使い方をする人はあまりいないかもしれませんが・・・。ちなみに、バイト(bのオプションを指定)として読み込まないとエラーを吐かれました。
with open('data/challenger_data.csv', 'rb') as cin: challenger_data_2 = np.genfromtxt(cin, skip_header=1, usecols=[1, 2], missing_values="NA", delimiter=",")
challenger_data_2
Output:
array([[ 66., 0.],
[ 70., 1.],
[ 69., 0.],
[ 80., nan],
[ 68., 0.],
[ 67., 0.],
[ 72., 0.],
[ 73., 0.],
[ 70., 0.],
[ 57., 1.],
[ 63., 1.],
[ 70., 1.],
[ 78., 0.],
[ 67., 0.],
[ 53., 1.],
[ 67., 0.],
[ 75., 0.],
[ 70., 0.],
[ 81., 0.],
[ 76., 0.],
[ 79., 0.],
[ 75., 1.],
[ 76., 0.],
[ 58., 1.],
[ 31., nan]])
skip_header引数
skip_header引数はファイルの上から何行を無視するかを指定する引数です。以下の例では、先頭から10行を無視するように指定したので、challenger_data_3はskip_header=1を指定したchallenger_dataより少ないレコードしか所持していません。
challenger_data_3 = np.genfromtxt("data/challenger_data.csv", skip_header=10, usecols=[1, 2], missing_values="NA", delimiter=",") challenger_data_3
Output:
array([[ 57., 1.],
[ 63., 1.],
[ 70., 1.],
[ 78., 0.],
[ 67., 0.],
[ 53., 1.],
[ 67., 0.],
[ 75., 0.],
[ 70., 0.],
[ 81., 0.],
[ 76., 0.],
[ 79., 0.],
[ 75., 1.],
[ 76., 0.],
[ 58., 1.],
[ 31., nan]])
usecols引数
usecols引数はどの列を使用するかを指定する引数です。例では、usecols=[1, 2]を指定しています。challenger_data.csvをみてみると、1列目には日付が入っていて、今回はそれを無視しています。
ちなみに、引数の順番通りにデータが渡されるらしく、以下のように列の順番を変えることもできます。
challenger_data_4 = np.genfromtxt("data/challenger_data.csv", skip_header=1, usecols=[2, 1], missing_values="NA", delimiter=",") challenger_data_4
Output:
array([[ 0., 66.],
[ 1., 70.],
[ 0., 69.],
[ nan, 80.],
[ 0., 68.],
[ 0., 67.],
[ 0., 72.],
[ 0., 73.],
[ 0., 70.],
[ 1., 57.],
[ 1., 63.],
[ 1., 70.],
[ 0., 78.],
[ 0., 67.],
[ 1., 53.],
[ 0., 67.],
[ 0., 75.],
[ 0., 70.],
[ 0., 81.],
[ 0., 76.],
[ 0., 79.],
[ 1., 75.],
[ 0., 76.],
[ 1., 58.],
[ nan, 31.]])
missing_values引数とfilling_values引数
missing_valuesは入力されたデータの中で、欠損値が何で表されているかを指定します。challenger_data.csvではNAという文字列で欠損値が表されているので、missing_values="NA"という指定になります。ただ、challenger_data.csvの最終行にはChallenger Accidentという文字列が入力されていましたが、その場合もnanに変換されていました。そこで、missing_valuesを指定せずに実行したら、同じ結果が得られました。もしかしたら、暗黙的な処理があり、ある程度の変換は行ってくれているかもしれません。
challenger_data_5 = np.genfromtxt("data/challenger_data.csv", skip_header=1, usecols=[1, 2], delimiter=",") challenger_data_5
Output:
array([[ 66., 0.],
[ 70., 1.],
[ 69., 0.],
[ 80., nan],
[ 68., 0.],
[ 67., 0.],
[ 72., 0.],
[ 73., 0.],
[ 70., 0.],
[ 57., 1.],
[ 63., 1.],
[ 70., 1.],
[ 78., 0.],
[ 67., 0.],
[ 53., 1.],
[ 67., 0.],
[ 75., 0.],
[ 70., 0.],
[ 81., 0.],
[ 76., 0.],
[ 79., 0.],
[ 75., 1.],
[ 76., 0.],
[ 58., 1.],
[ 31., nan]])
missing_values引数により欠損値だと判断されたデータはfilling_values引数で指定したデータが使用されるようです。今回は指定されていませんが、こちらも暗黙的な処理が裏で走るようです。
指定できるデータ型はその列のデータ型のみのようです。無効なデータ型をfilling_valuesに渡すとエラーになります。
challenger_data_6 = np.genfromtxt("data/challenger_data.csv", skip_header=1, usecols=[1, 2], missing_values='NA', filling_values='missing_value', delimiter=",") challenger_data_6
Error:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-9-3069a5bf63ec> in <module>
2 skip_header=1, usecols=[1, 2],
3 missing_values='NA', filling_values='missing_value',
----> 4 delimiter=",")
5 challenger_data_6
~\Anaconda3\lib\site-packages\numpy\lib\npyio.py in genfromtxt(fname, dtype, comments, delimiter, skip_header, skip_footer, converters, missing_values, filling_values, usecols, names, excludelist, deletechars, replace_space, autostrip, case_sensitive, defaultfmt, unpack, usemask, loose, invalid_raise, max_rows)
1868 dtype = np.dtype(ttype)
1869 #
-> 1870 output = np.array(data, dtype)
1871 if usemask:
1872 if dtype.names:
ValueError: could not convert string to float: 'missing_value'
float型なら問題なく設定できます。
challenger_data_6 = np.genfromtxt("data/challenger_data.csv", skip_header=1, usecols=[1, 2], missing_values='NA', filling_values=1000., delimiter=",") challenger_data_6
Output:
array([[ 66., 0.],
[ 70., 1.],
[ 69., 0.],
[ 80., 1000.],
[ 68., 0.],
[ 67., 0.],
[ 72., 0.],
[ 73., 0.],
[ 70., 0.],
[ 57., 1.],
[ 63., 1.],
[ 70., 1.],
[ 78., 0.],
[ 67., 0.],
[ 53., 1.],
[ 67., 0.],
[ 75., 0.],
[ 70., 0.],
[ 81., 0.],
[ 76., 0.],
[ 79., 0.],
[ 75., 1.],
[ 76., 0.],
[ 58., 1.],
[ 31., 1000.]])
delimiter引数
これは、データの区切り文字を表しています。例ではコンマ(,)により区切られていました。
また、自然数か、自然数のリストを渡すと、その長さで区切られたデータが格納されるようです。使い道あるのかな・・・?
challenger_data_7 = np.genfromtxt("data/challenger_data.csv", skip_header=1, missing_values="NA",dtype=str, delimiter=[8, 8, 8, 8]) challenger_data_7
Output:
array([['04/12/19', '81,66,0\n', '', ''],
['11/12/19', '81,70,1\n', '', ''],
['3/22/82,', '69,0\n', '', ''],
['6/27/82,', '80,NA\n', '', ''],
['01/11/19', '82,68,0\n', '', ''],
['04/04/19', '83,67,0\n', '', ''],
['6/18/83,', '72,0\n', '', ''],
['8/30/83,', '73,0\n', '', ''],
['11/28/83', ',70,0\n', '', ''],
['02/03/19', '84,57,1\n', '', ''],
['04/06/19', '84,63,1\n', '', ''],
['8/30/84,', '70,1\n', '', ''],
['10/05/19', '84,78,0\n', '', ''],
['11/08/19', '84,67,0\n', '', ''],
['1/24/85,', '53,1\n', '', ''],
['04/12/19', '85,67,0\n', '', ''],
['4/29/85,', '75,0\n', '', ''],
['6/17/85,', '70,0\n', '', ''],
['7/29/85,', '81,0\n', '', ''],
['8/27/85,', '76,0\n', '', ''],
['10/03/19', '85,79,0\n', '', ''],
['10/30/85', ',75,1\n', '', ''],
['11/26/85', ',76,0\n', '', ''],
['01/12/19', '86,58,1\n', '', ''],
['1/28/86,', '31,Chall', 'enger Ac', 'cident\n']],
dtype='<U8')
解説は以上です。記事を書いているときにわからないことがたくさん出てきたので、まだまだ知らなければいけないことはたくさんあるなぁと感じます。
また、記事を書いている途中で、Numpyのデータ型について知っていないといけないなぁとも思いました。難しいです。
Pythonで体験するベイズ推論 PyMCによるMCMC入門 第1章 写経
始めに
この記事では、Pythonで体験するベイズ推論 PyMCによるMCMC入門
の第1章を写経した記録をまとめます。実行はJupyter Labにて実行しています。
写経は以下のようにまとめました。
写経ではありますが、コードを実行しやすいように関数を作成し、コードの再実行がやりやすいようにしてあります。
また、より良いと思われるコードを考えた場合は書き換えてコメントを添えるようにし、変更点をなるべく明示するようにしてあります。
個人的に気になった点のメモを残すようにしてあります。同じような疑問を持った方の助けになれば幸いです。
Pythonで体験するベイズ推論 PyMCによるMCMC入門を読んでいている際のちょっとした正誤表代わりになればと思います。
1.2 ベイズ推論の枠組み
1.2.2 例題:司書か農家か?
メモ
IPython.core.pylabtoolsはIpythonのAPIの一つで、Pylab (matplotlib) の操作をサポートしてくれる関数がまとめられています。インポートしたfigsize関数はデフォルトの画像サイズを設定してくれる関数です。
figsize(12.5, 4)は
matplotlib.rcParams['figure.figsize'] = [25, 8]
を実行することでできるそう。
from IPython.core.pylabtools import figsize import numpy as np from matplotlib import pyplot as plt
%matplotlib inline figsize(25, 8) # テキストの値だと、画像が小さかったので、2倍のサイズを指定した。 plt.rcParams['font.size'] = 25 # フォントのサイズを大きくする。
colors = ["#348ABD", "#A60628"] prior = [1 / 21., 20 / 21.] posterior = [0.087, 1 - 0.087] plt.bar([0, .7], prior, alpha=0.70, width=0.25, color=colors[0], label="prior distribution", lw="3", edgecolor=colors[0]) # テキストでは、edgecolor="#348ABD"と書いてあったが、これはcolors[0]のことなので書き直した。 plt.bar([0 + 0.25, .7 + 0.25], posterior, alpha=0.7, width=0.25, color=colors[1], label="posterior distribution", lw="3", edgecolor=colors[1]) # テキストでは、edgecolor="#A60628"と書いてあったが、これはcolors[1]のことなので書き直した。 plt.xticks([0.20, 0.95], ["Librarian", "Farmer"]) plt.xlabel("Probability") plt.legend(loc="upper left") plt.title("Prior and posterior probabilities of Steve's occupation")
Text(0.5, 1.0, "Prior and posterior probabilities of Steve's occupation")

1.3 確率分布
1.3.1 離散の場合
import scipy.stats as stats
def plot_two_poi_dist(lambda_0=1.5, lambda_1=4.25): poi = stats.poisson colors = ["#348ABD", "#A60628"] a = np.arange(16) plt.bar(a, poi.pmf(a, lambda_0), color=colors[0], label=f"$\lambda = {lambda_0}$", # テキストだと%によりlambda_[0]の値を文字列の中に埋め込んでいたが、fを使えばよいだろう。%を使うことに何か利点があるのかな? alpha=0.6, edgecolor=colors[0], lw=3) #alphaはバーの透明度。0だと透明(見えない)、1だと透過なし。 #lwはエッジの幅のようだ。テキストはなぜか"3"のように文字列を指定していたが、整数を渡せばいいと思う。エラーにならないのは、整数への変換が内部で起っているのかな? plt.bar(a, poi.pmf(a, lambda_1), color=colors[1], label=f"$\lambda = {lambda_1}$", alpha=0.60, edgecolor=colors[1], lw=3) plt.xticks(a + 0.4, a) plt.legend() plt.ylabel("Probability of $k$") plt.xlabel("$k$") plt.title("Probability mass function of a Poisson random variable, differing $\lambda$ values")
plot_two_poi_dist()

メモ
モジュールscipy.statsは種々の確率質量関数と確率密度関数を生成するクラスをまとめたもののようです。
参考:Statistics (scipy.stats)、Statistical functions (scipy.stats)
たとえば、上のコードにあるように
poi = stats.poisson
type(poi)
scipy.stats._discrete_distns.poisson_gen
により作成されたクラスpoisson_genのインスタンスpoiは、メソッドpmfによりポアソン分布の確率質量関数の値を計算することができます。
poi.pmf(3, 4) # 平均が4であるポアソン分布の確率質量関数がx=3においてとる値を計算する。
0.19536681481316454
また、平均や分散も計算してくれるようです。
poi.stats(4)
(array(4.0), array(4.0))
当たり前のように、ポアソン分布に従う確率変数を生成することもできます。メソッドrvsを使います。
poi.rvs(4, size=10)
array([ 7, 7, 5, 1, 10, 7, 4, 8, 1, 6])
ちなみに、平均を指定してインスタンスを作成することができます。
poi_4 = stats.poisson(4)
poi_4.pmf(3)
0.19536681481316454
このようにして作成されたインスタンスはfrozen objectなどと呼ばれるようです。
Alternatively, the distribution object can be called (as a function) to fix the shape and location. This returns a “frozen” RV object holding the given parameters fixed.
平均を指定せずに作成したpoiとは異なるクラスになるようです。
type(poi_4)
scipy.stats._distn_infrastructure.rv_frozen
1.3.2 連続の場合
def plot_two_expo_dist(lambda_=[0.5, 4]): a = np.linspace(0, 4, 100) expo = stats.expon colors = ["#348ABD", "#A60628"] for l, c in zip(lambda_, colors): plt.plot(a, expo.pdf(a, scale=1. / l), lw=3, color=c, label=f"$\lambda = {l}") plt.fill_between(a, expo.pdf(a, scale=1. / l), color=c, alpha=.33) plt.legend() plt.ylabel("Probability density function at $z$") plt.xlabel("$z$") plt.ylim(0, 1.2) plt.title("Probability density function of an exponential " "random variable, differing $\lambda$ values")
plot_two_expo_dist()

メモ
fill_betweenは二つの曲線の間を塗りつぶす関数のようです。
参考:matplotlib.pyplot.fill_between
次の関数のように、二つの曲線(expo.pdf(a, scale=1. / lambda_[0])とexpo.pdf(a, scale=1. / lambda_[1]))を指定すると、その間を塗りつぶしてくれる。
def plot_two_expo_dist2(lambda_=[0.5, 4]): a = np.linspace(0, 4, 100) expo = stats.expon colors = ["#348ABD", "#A60628"] for l, c in zip(lambda_, colors): plt.plot(a, expo.pdf(a, scale=1. / l), lw=3, color=c, label=f"$\lambda = {l}$") plt.fill_between(a, expo.pdf(a, scale=1. / lambda_[0]), expo.pdf(a, scale=1. / lambda_[1]), color="#00d000", alpha=.33) plt.legend() plt.ylabel("Probability density function at $z$") plt.xlabel("$z$") plt.ylim(0, 1.2) plt.title("Probability density function of an exponential " "random variable, differing $\lambda$ values")
plot_two_expo_dist2()

1.4 コンピュータにベイズ推論をさせるには
1.4.1 例題:メッセージ数に変化はあるか?
from os import makedirs makedirs("data", exist_ok=True)
from urllib.request import urlretrieve urlretrieve("https://git.io/vXTVC", "data/txtdata.csv")
('data/txtdata.csv', <http.client.HTTPMessage at 0x2e155deb8d0>)
count_data = np.loadtxt("data/txtdata.csv")
def plot_uss_txt_hab(count_data=count_data): n_count_data = len(count_data) plt.bar(np.arange(n_count_data), count_data, color="#348ABD") plt.xlabel("Time (days)") plt.ylabel("Text messages received") plt.title("Did the user's texting habits change over time?") plt.xlim(0, n_count_data)
plot_uss_txt_hab()

1.4.2 一撃必殺:PyMC
import pymc as pm
alpha = 1.0 / count_data.mean() lambda_1 = pm.Exponential("lambda_1", alpha) lambda_2 = pm.Exponential("lambda_2", alpha) n_count_data = len(count_data) tau = pm.DiscreteUniform("tau", lower=0, upper=n_count_data) print(f"Random output: {tau.random()} {tau.random()} {tau.random()}")
Random output: 10 48 50
@pm.deterministic def lambda_(tau=tau, lambda_1=lambda_1, lambda_2=lambda_2): out = np.zeros(n_count_data) out[:tau] = lambda_1 out[tau:] = lambda_2 return out
observation = pm.Poisson("obs", lambda_, value=count_data, observed=True) model = pm.Model([observation, lambda_1, lambda_2, tau])
mcmc = pm.MCMC(model) mcmc.sample(40000, 10000)
[-----------------100%-----------------] 40000 of 40000 complete in 6.6 sec
def plot_post_dist(mcmc=mcmc): figsize(29, 20) lambda_1_samples = mcmc.trace('lambda_1')[:] lambda_2_samples = mcmc.trace('lambda_2')[:] tau_samples = mcmc.trace('tau')[:] ax = plt.subplot(311) ax.set_autoscaley_on(False) plt.hist(lambda_1_samples, histtype='stepfilled', bins=30, alpha=0.85, color="#A60628", density=True, # normedという引数はどうやらMatplotlib 3.1では削除されてしまうらしいのでdensityを使用した。 label="posterior of $\lambda_1$") plt.legend(loc="upper left") plt.title("Posterior distributions of the parameters " r"$\lambda_1, \lambda_2, \tau$") plt.xlim([15, 30]) plt.xlabel("$\lambda_1$ value") plt.ylabel("Density") ax = plt.subplot(312) ax.set_autoscaley_on(False) plt.hist(lambda_2_samples, histtype='stepfilled', bins=30, alpha=0.85, color="#7A68A6", density=True, # normedという引数はどうやらMatplotlib 3.1では削除されてしまうらしいのでdensityを使用した。 label="posterior of $\lambda_2$") plt.legend(loc="upper left") plt.xlim([15, 30]) plt.xlabel("$\lambda_2$ value") plt.ylabel("Density") plt.subplot(313) w = 1.0 / tau_samples.shape[0] * np.ones_like(tau_samples) plt.hist(tau_samples, bins=n_count_data, alpha=1, label=r"posterior of $\tau$", color="#467821", weights=w, rwidth=2.) plt.xticks(np.arange(n_count_data)) plt.legend(loc="upper left") plt.ylim([0, .75]) plt.xlim([35, len(count_data) - 20]) plt.xlabel(r"$\tau$ (in days)") plt.ylabel("Probability")
plot_post_dist()
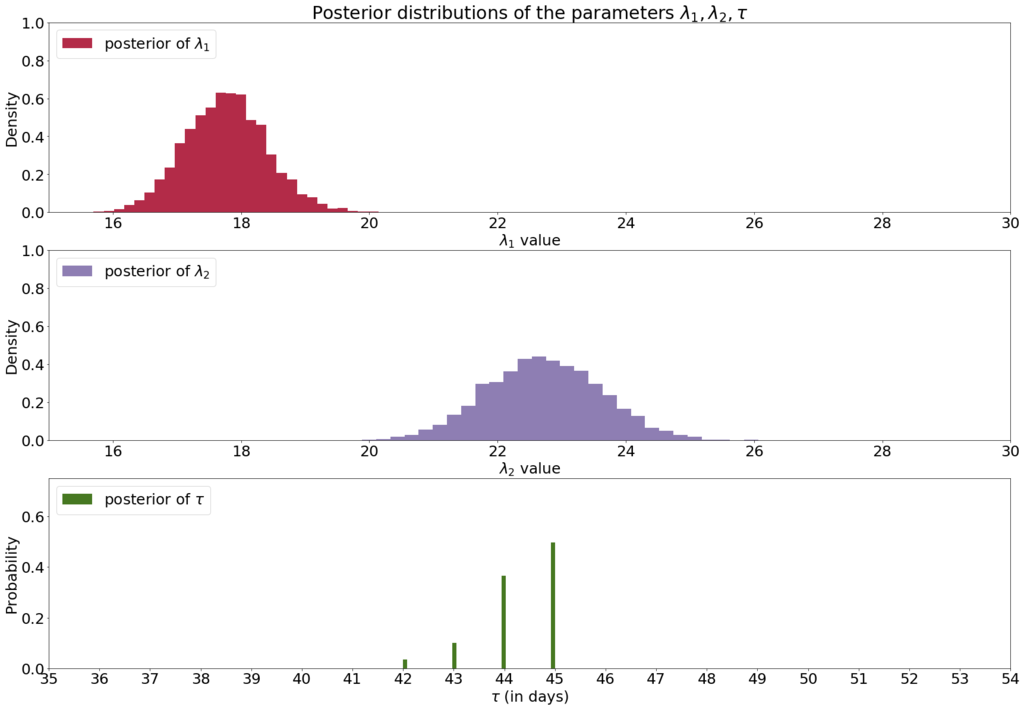
def plot_expc_txt(mcmc=mcmc, count_data=count_data, n_count_data=n_count_data): figsize(25, 8) lambda_1_samples = mcmc.trace('lambda_1')[:] lambda_2_samples = mcmc.trace('lambda_2')[:] tau_samples = mcmc.trace('tau')[:] N = tau_samples.shape[0] expected_texts_per_day = np.zeros(n_count_data) for day in range(n_count_data): ix = day < tau_samples expected_texts_per_day[day] = (lambda_1_samples[ix].sum() + lambda_2_samples[~ix].sum()) / N plt.plot(range(n_count_data), expected_texts_per_day, lw=4, color="#E24A33", label="Expected number of text messages received") plt.xlim(0, n_count_data) plt.ylim(0, 60) plt.xlabel("Day") plt.ylabel("Number of text messages") plt.title("Number of text messages received versus expected number received") plt.bar(np.arange(len(count_data)), count_data, color="#348ABD", alpha=0.65, label="Observed text messages per day") plt.legend(loc="upper left") print(expected_texts_per_day)
plot_expc_txt()
[ 17.76436598 17.76436598 17.76436598 17.76436598 17.76436598
17.76436598 17.76436598 17.76436598 17.76436598 17.76436598
17.76436598 17.76436598 17.76436598 17.76436598 17.76436598
17.76436598 17.76436598 17.76436598 17.76436598 17.76436598
17.76436598 17.76436598 17.76436598 17.76436598 17.76436598
17.76436598 17.76436598 17.76436598 17.76436598 17.76436598
17.76436598 17.76436598 17.76436598 17.76436598 17.76436598
17.76436598 17.76436598 17.76436598 17.76436598 17.76451967
17.76484233 17.7669707 17.91943442 18.38499061 20.19507103
22.72249711 22.72249711 22.72249711 22.72249711 22.72249711
22.72249711 22.72249711 22.72249711 22.72249711 22.72249711
22.72249711 22.72249711 22.72249711 22.72249711 22.72249711
22.72249711 22.72249711 22.72249711 22.72249711 22.72249711
22.72249711 22.72249711 22.72249711 22.72249711 22.72249711
22.72249711 22.72249711 22.72249711 22.72249711]

1.5 おわりに
付録
実際に二つのλは異なっているのか?
lambda_1_samples = mcmc.trace('lambda_1')[:] lambda_2_samples = mcmc.trace('lambda_2')[:] tau_samples = mcmc.trace('tau')[:]
print(lambda_1_samples < lambda_2_samples)
[ True True True ..., True True True]
print((lambda_1_samples < lambda_2_samples).sum()) print(lambda_1_samples.shape[0])
30000
30000
どうやら、僕がやったシミュレーションではlambda_2_samplesの全ての要素がlambda_1_samplesの全ての要素より大きかったようです。もちろん確率も1。
print((lambda_1_samples < lambda_2_samples).mean())
1.0
for d in [1, 2, 5, 10]: v = (abs(lambda_1_samples - lambda_2_samples) >= d).mean() print(f"What is the probability the difference is larger than {d}? {round(v, 2)}")
What is the probability the difference is larger than 1? 1.0
What is the probability the difference is larger than 2? 1.0
What is the probability the difference is larger than 5? 0.49
What is the probability the difference is larger than 10? 0.0
二つの変化点への拡張
def plot_post_dist(count_data=count_data): figsize(29, 20) alpha = 1.0 / count_data.mean() lambda_1 = pm.Exponential("lambda_1", alpha) lambda_2 = pm.Exponential("lambda_2", alpha) lambda_3 = pm.Exponential("lambda_3", alpha) n_count_data = len(count_data) tau_1 = pm.DiscreteUniform("tau_1", lower=0, upper=n_count_data - 1) tau_2 = pm.DiscreteUniform("tau_2", lower=tau_1, upper=n_count_data) @pm.deterministic def lambda_(tau_1=tau_1, tau_2=tau_2, lambda_1=lambda_1, lambda_2=lambda_2, lambda_3=lambda_3): out = np.zeros(n_count_data) out[:tau_1] = lambda_1 out[tau_1:tau_2] = lambda_2 out[tau_2:] = lambda_3 return out observation = pm.Poisson("obs", lambda_, value=count_data, observed=True) model = pm.Model([observation, lambda_1, lambda_2, lambda_3, tau_1, tau_2]) mcmc = pm.MCMC(model) mcmc.sample(40000, 10000) lambda_1_samples = mcmc.trace('lambda_1')[:] lambda_2_samples = mcmc.trace('lambda_2')[:] lambda_3_samples = mcmc.trace('lambda_3')[:] tau1_samples = mcmc.trace('tau_1')[:] tau2_samples = mcmc.trace('tau_2')[:] # lambda_達をプロットするときにxlimを設定するのに使う lambda_min = min([min(lambda_1_samples), min(lambda_2_samples), min(lambda_3_samples)]) lambda_max = max([max(lambda_1_samples), max(lambda_2_samples), max(lambda_3_samples)]) ax = plt.subplot(511) ax.set_autoscaley_on(False) plt.hist(lambda_1_samples, histtype='stepfilled', bins=30, alpha=0.85, color="#A60628", density=True, # normedという引数はどうやらMatplotlib 3.1では削除されてしまうらしいのでdensityを使用した。 label="posterior of $\lambda_1$") plt.legend(loc="upper left") plt.title("Posterior distributions of the five unknown " "parameters in the extended text-message model") plt.xlim([lambda_min - 1, lambda_max + 1]) plt.xlabel("$\lambda_1$ value") plt.ylabel("Density") plt.subplot(512) ax.set_autoscaley_on(False) plt.hist(lambda_2_samples, histtype='stepfilled', bins=30, alpha=0.85, color="#7A68A6", density=True, # normedという引数はどうやらMatplotlib 3.1では削除されてしまうらしいのでdensityを使用した。 label="posterior of $\lambda_2$") plt.legend(loc="upper left") plt.xlim([lambda_min - 1, lambda_max + 1]) plt.xlabel("$\lambda_2$ value") plt.ylabel("Density") ax = plt.subplot(513) ax.set_autoscaley_on(False) plt.hist(lambda_3_samples, histtype='stepfilled', bins=30, alpha=0.85, color="#7A68A6", density=True, # normedという引数はどうやらMatplotlib 3.1では削除されてしまうらしいのでdensityを使用した。 label="posterior of $\lambda_3$") plt.legend(loc="upper left") plt.xlim([lambda_min - 1, lambda_max + 1]) plt.xlabel("$\lambda_3$ value") plt.ylabel("Density") ax = plt.subplot(514) w = 1.0 / tau1_samples.shape[0] * np.ones_like(tau1_samples) plt.hist(tau1_samples, bins=n_count_data, alpha=1, label=r"posterior of $\tau_1$", color="#467821", weights=w, rwidth=2.) plt.xticks(np.arange(n_count_data)) plt.legend(loc="upper left") plt.ylim([0, .75]) plt.xlim([min(tau1_samples) - 1, max(tau2_samples) + 1]) # テキストだと、35日目から54日目までを固定してプロットしてるが、 #これだとmcmc.sampleの結果によっては範囲外にプロットされてしまうのでこのように書き換えた。 plt.xlabel("Day") plt.ylabel("Probability") ax = plt.subplot(515) w = 1.0 / tau2_samples.shape[0] * np.ones_like(tau2_samples) plt.hist(tau2_samples, bins=n_count_data, alpha=1, label=r"posterior of $\tau_2$", color="#467821", weights=w, rwidth=2.) plt.xticks(np.arange(n_count_data)) plt.legend(loc="upper left") plt.ylim([0, .75]) plt.xlim([min(tau1_samples) - 1, max(tau2_samples) + 1]) # テキストだと、35日目から54日目までを固定してプロットしてるが、 #これだとmcmc.sampleの結果によっては範囲外にプロットされてしまうのでこのように書き換えた。 plt.xlabel("Day") plt.ylabel("Probability")
mcmc.sample(40000, 10000)の結果によっては、テキストの結果と大分違う場合があるようです。
plot_post_dist()
[-----------------100%-----------------] 40000 of 40000 complete in 10.1 sec

plot_post_dist()
[-----------------100%-----------------] 40000 of 40000 complete in 14.5 sec

plot_post_dist()
[-----------------100%-----------------] 40000 of 40000 complete in 10.1 sec

今回の写経は以上です。 ここまで読んでいただきありがとうございました。


